Ann Lab Med.
2012 Jan;32(1):50-56. 10.3343/alm.2012.32.1.50.
Comparison of Modified Multiple-locus Variable-number Tandem-repeat Fingerprinting with Pulsed-field Gel Electrophoresis for Typing Clinical Isolates of Staphylococcus aureus
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul, Korea. euichong@snu.ac.kr
- 2Department of Laboratory Medicine, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 3Department of Laboratory Medicine, Pusan National University School of Medicine, Busan, Korea.
- 4Department of Laboratory Medicine, National Medical Center, Seoul, Korea.
- KMID: 1781438
- DOI: http://doi.org/10.3343/alm.2012.32.1.50
Abstract
- BACKGROUND
Multiple-locus variable-number tandem-repeat fingerprinting (MLVF) is based on multiplex PCR, utilizing variable number tandem repeat. Our goal was to compare the performance of MLVF in distinguishing clinical Staphylococcus aureus isolates with that of pulsed-field gel electrophoresis (PFGE), which has traditionally been the gold standard.
METHODS
Sixty-three clinically significant S. aureus isolates were tested using both PFGE and MLVF. Multiplex PCR for MLVF was performed using PCR primers for clfA, clfB, sdrCDE, sspA, and spa. PFGE was performed with genomic DNA fragments generated by SmaI endonuclease digestion. Banding patterns of MLVF or PFGE were analyzed using InfoQuestFP software.
RESULTS
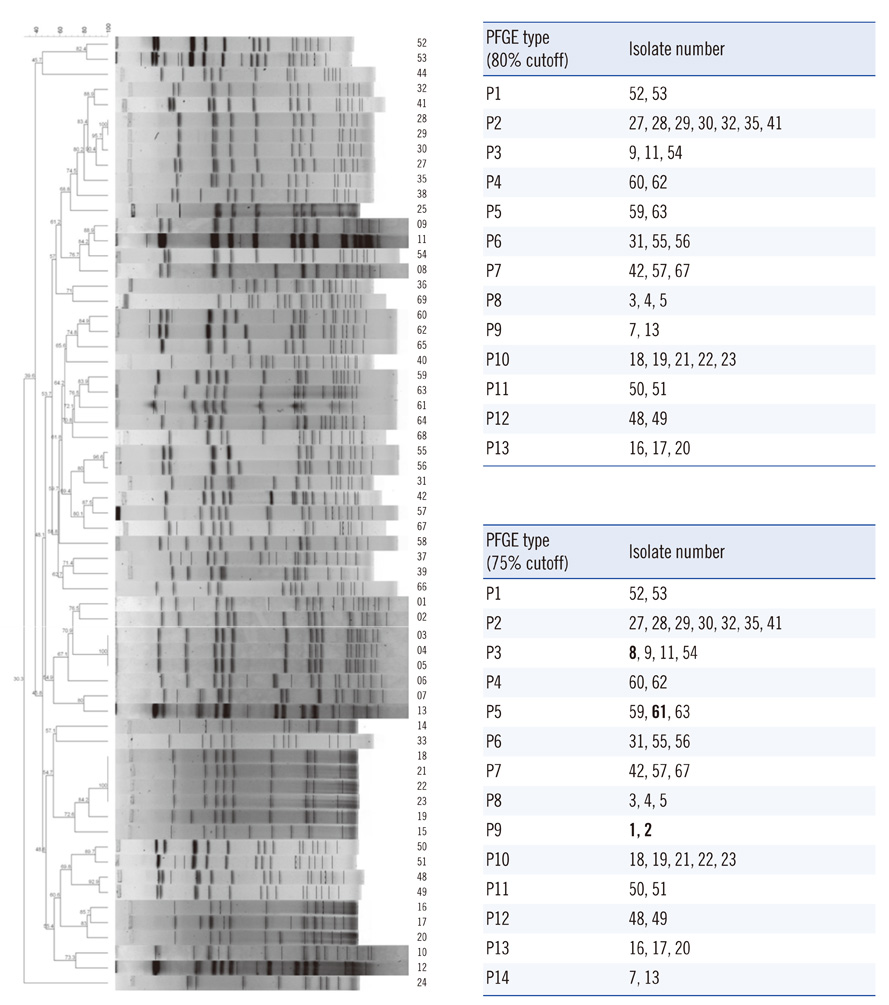
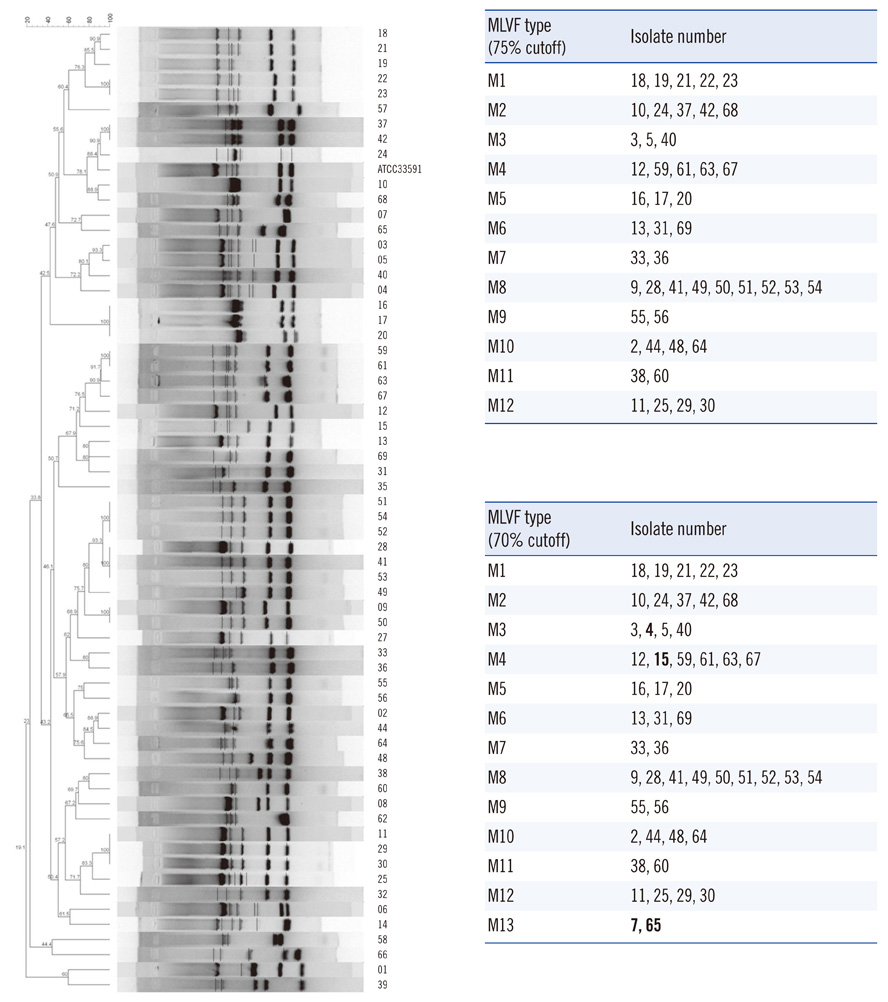
The hands-on time of our modified method was about 3 h, on average, for each of 18 isolates. PFGE (80% cutoff) or MLVF (75% cutoff) separated all of the 63 isolates into 13 and 12 types, respectively. Three types generated by PFGE were identical to those generated by MLVF. PFGE and MLVF yielded similar Simpson's diversity indices, indicating similar discriminatory power. The overall concordance between PFGE and MLVF was low, as represented by adjusted Rand indices (0.266-0.278). PFGE predicted MLVF type better than MLVF predicted PFGE type, as reflected by Wallace coefficients (PFGE cutoff 80% vs. MLVF cutoff 75%, 0.389 vs. 0.233). Analysis of the relationship between a pair of isolates showed 91.0% concordance between the PFGE (80% cutoff) and MLVF (75% cutoff).
CONCLUSIONS
Our simple, low-cost, modified MLVF protocol can effectively discriminate between S. aureus clinical isolates. MLVF can replace PFGE for the hospital infection control of S. aureus.
Keyword
MeSH Terms
-
Bacterial Typing Techniques/*methods
*DNA Fingerprinting
DNA, Bacterial/analysis
*Electrophoresis, Gel, Pulsed-Field
Genotype
Humans
Methicillin-Resistant Staphylococcus aureus/classification/genetics/isolation & purification
Multiplex Polymerase Chain Reaction
Staphylococcal Infections/*microbiology
Staphylococcus aureus/*classification/*genetics/isolation & purification
Figure
Reference
-
1. Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004. 364:703–705.2. Wertheim HF, van Leeuwen WB, Snijders S, Vos MC, Voss A, Vandenbroucke-Grauls CM, et al. Associations between Staphylococcus aureus genotype, infection, and in-hospital mortality: a nested case-control study. J Infect Dis. 2005. 192:1196–1200.3. Cosgrove SE. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis. 2006. 42:Suppl 2. S82–S89.
Article4. Struelens MJ, Deplano A, Godard C, Maes N, Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992. 30:2599–2605.5. Tenover FC, Vaughn RR, McDougal LK, Fosheim GE, McGowan JE Jr. Multiple-locus variable-number tandem-repeat assay analysis of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007. 45:2215–2219.6. Sabat A, Krzyszton-Russjan J, Strzalka W, Filipek R, Kosowska K, Hryniewicz W, et al. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J Clin Microbiol. 2003. 41:1801–1804.7. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, et al. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J Clin Microbiol. 2005. 43:3095–3100.8. Moser SA, Box MJ, Patel M, Amaya M, Schelonka R, Waites KB. Multiple-locus variable-number tandem-repeat analysis of meticillin-resistant Staphylococcus aureus discriminates within USA pulsed-field gel electrophoresis types. J Hosp Infect. 2009. 71:333–339.9. Horan TC, Andrus M, Dudeck MA. CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008. 36:309–332.
Article10. Chung M, de Lencastre H, Matthews P, Tomasz A, Adamsson I, Aires de Sousa M, et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist. 2000. 6:189–198.11. Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Sousa H, Almeida JS, et al. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol. 2006. 44:2524–2532.12. Robinson DA, Hollingshead SK, Musser JM, Parkinson AJ, Briles DE, Crain MJ. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J Mol Evol. 1998. 47:222–229.13. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003. 41:5113–5120.14. Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, et al. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One. 2009. 4:e5082.15. Sabat A, Malachowa N, Miedzobrodzki J, Hryniewicz W. Comparison of PCR-based methods for typing Staphylococcus aureus isolates. J Clin Microbiol. 2006. 44:3804–3807.16. Holmes A, Edwards GF, Girvan EK, Hannant W, Danial J, Fitzgerald JR, et al. Comparison of two multilocus variable-number tandem-repeat methods and pulsed-field gel electrophoresis for differentiating highly clonal methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 2010. 48:3600–3607.17. Rasschaert G, Vanderhaeghen W, Dewaele I, Janez N, Huijsdens X, Butaye P, et al. Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398. J Clin Microbiol. 2009. 47:3313–3322.18. David MZ, Boyle-Vavra S, Zychowski DL, Daum RS. Methicillin-susceptible Staphylococcus aureus as a predominantly healthcare-associated pathogen: a possible reversal of roles? PLoS One. 2011. 6:e18217.19. Bio-Rad laboratories, Inc. InfoQuest FP software user guide version 4.5. 2006. Hercules, CA: Bio-Rad laboratories, Inc.;179–180.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular Typing in Public Health Laboratories: From an Academic Indulgence to an Infection Control Imperative
- Molecular Typing of
Mycobacterium intracellulare Using Pulsed-Field Gel Electrophoresis, Variable-Number Tandem-Repeat Analysis, Mycobacteria Interspersed Repetitive-Unit-Variable-Number Tandem Repeat Typing, and Multilocus Sequence Typing: Molecular Characterization and Comparison of Each Typing Methods - Multilocus Sequence Typing for Candida albicans Isolates from Candidemic Patients: Comparison with Southern Blot Hybridization and Pulsed-field Gel Electrophoresis Analysis
- Typing and Antimicrobial Susceptibilities of Methicillin Resistant Staphylococcus aureus (MRSA) Strains Isolated in a Hospital in Korea
- Molecular Epidemiologic Analysis of Methicillin-Resistant Staphylococcus aureus using Pulsed-Field Gel Electrophoresis