Ann Lab Med.
2014 Jan;34(1):51-55. 10.3343/alm.2014.34.1.51.
Assessment of the Quantitative Ability of AdvanSure TB/NTM Real-Time PCR in Respiratory Specimens by Comparison with Phenotypic Methods
- Affiliations
-
- 1Department of Laboratory Medicine, School of Medicine, The Catholic University of Korea, Seoul, Korea. yjpk@catholic.ac.kr
- 2Department of Biostatistics, School of Medicine, The Catholic University of Korea, Seoul, Korea.
- KMID: 1781360
- DOI: http://doi.org/10.3343/alm.2014.34.1.51
Abstract
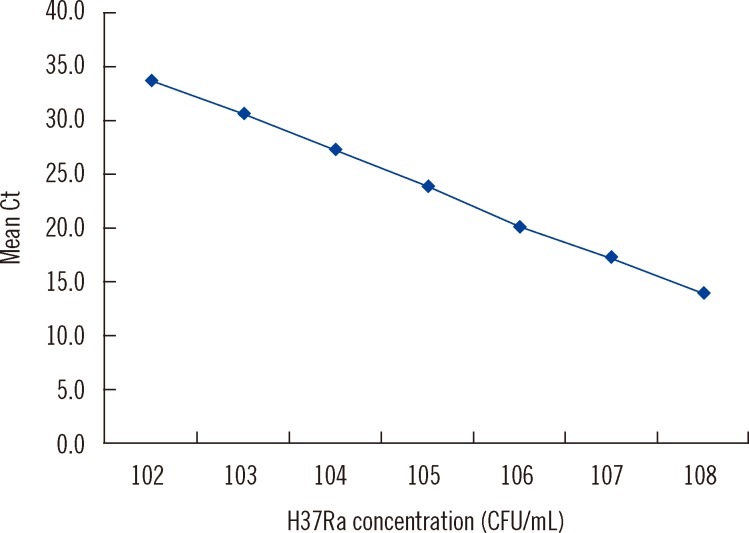
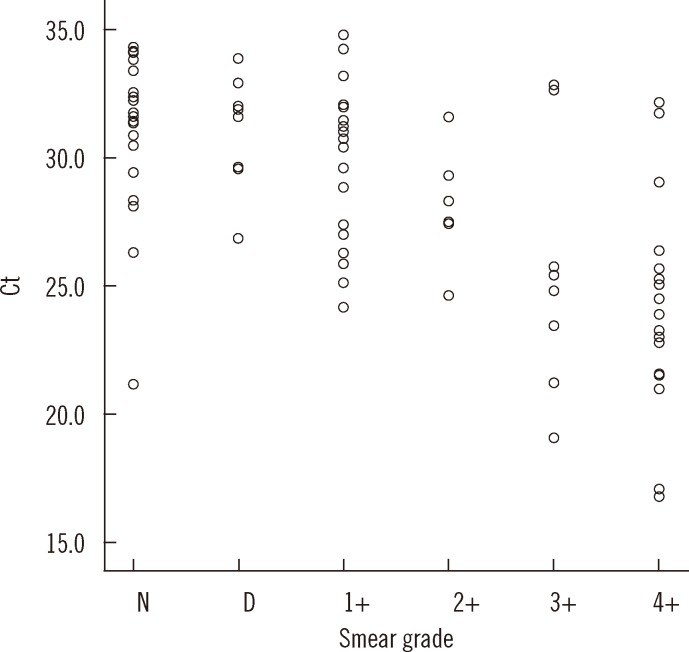
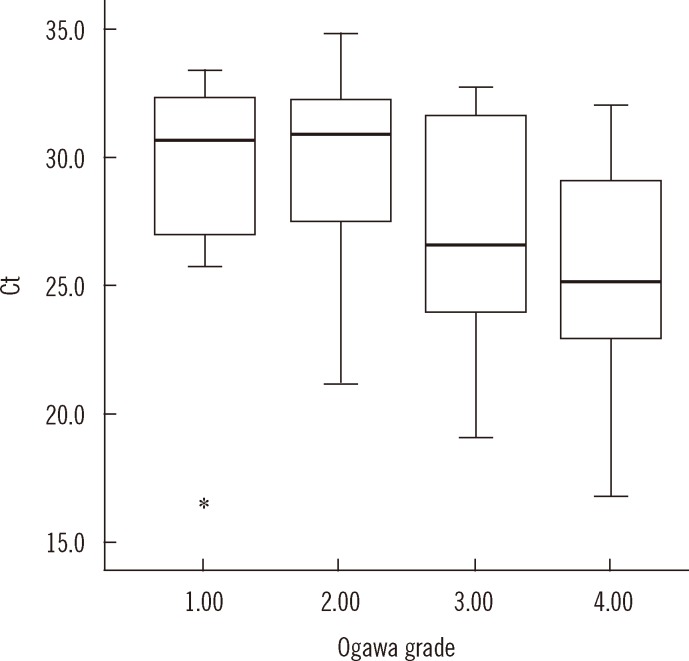
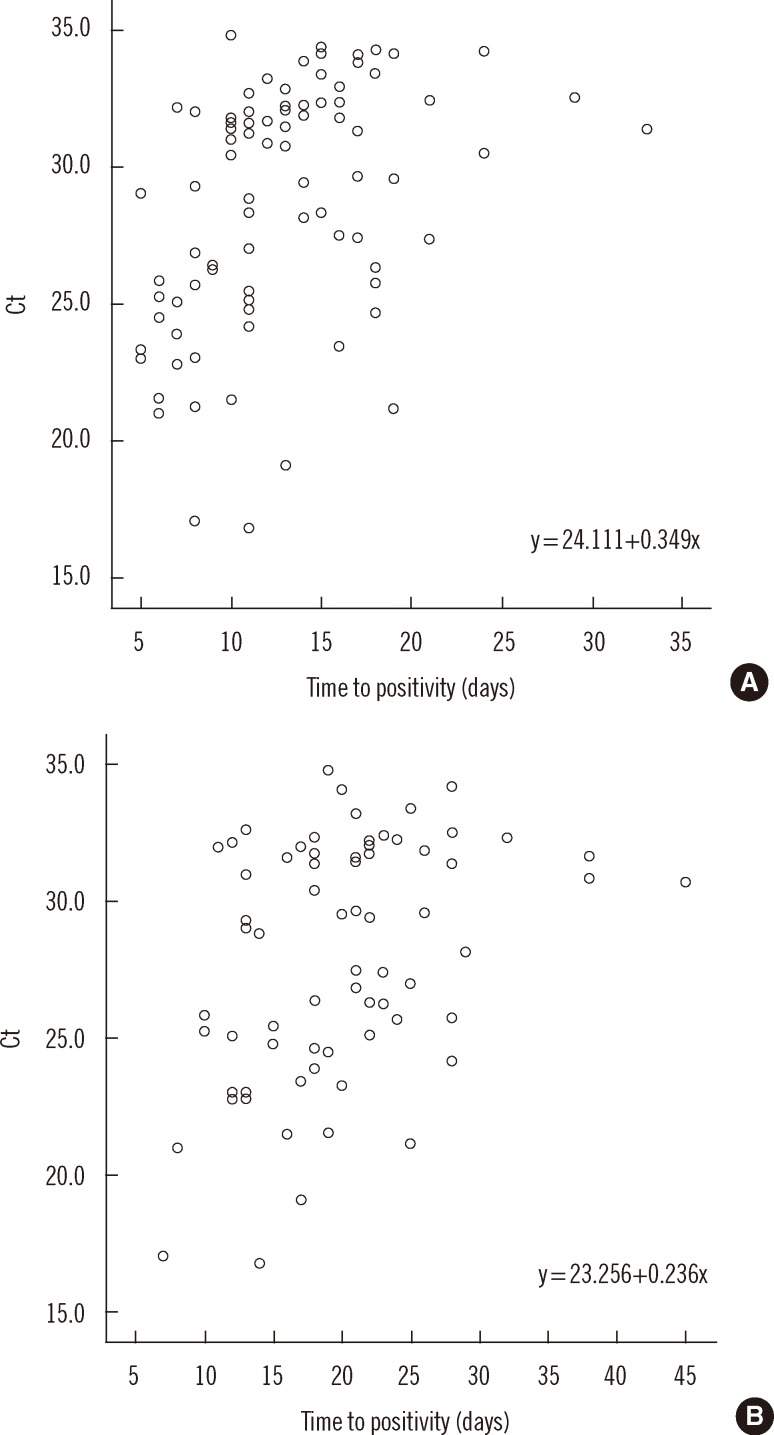
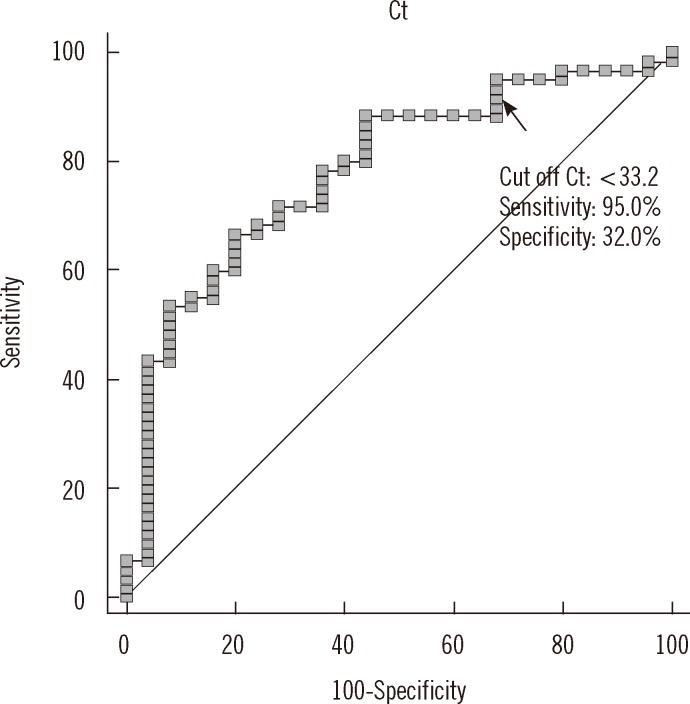
- Accurate quantification of mycobacterial load is important to evaluate disease severity and to monitor the course of treatment in tuberculosis (TB). We evaluated the quantitative capability of the AdvanSure TB/NTM real-time PCR kit (LG Life Science, Korea) to determine the cycle threshold (Ct) for mycobacterial burden. We retrospectively analyzed data from 108 patients whose respiratory specimens (sputums and bronchoalveolar lavage fluids) were positive for Mycobacterium tuberculosis complex (85 culture-positive and 23 culture-negative specimens). We compared Ct values with grades of acid-fast bacilli (AFB) staining, semi-quantitative colony count on solid medium, and time to positivity (TTP) in liquid and solid media. We also investigated the cutoff Ct value for predicting stain-positive status. Ct value showed significant reverse correlation with AFB staining grade (r(s)=-0.635, P<0.01). Ct value significantly decreased as the semi-quantitative counts on the solid medium increased (P<0.001), and the mean Ct value of each of the groups 1+, 2+, 3+, and 4+ were 29.0, 30.0, 27.1, and 25.5, respectively. A weak correlation between Ct value and TTP in liquid and solid media was observed (r(s)=0.468 and 0.365, respectively). A cutoff Ct value of <33.2 best predicted stain positivity, with a sensitivity of 95.0% and a specificity of 32.0%. Our findings suggest the potential use of AdvanSure TB/NTM real-time PCR kit for quantitatively determining bacterial burden, albeit with some enhancements.
MeSH Terms
-
Area Under Curve
Bronchoalveolar Lavage Fluid/microbiology
DNA, Bacterial/*analysis
Humans
Mycobacterium tuberculosis/genetics/*isolation & purification
Phenotype
ROC Curve
Reagent Kits, Diagnostic
*Real-Time Polymerase Chain Reaction
Retrospective Studies
Sensitivity and Specificity
Sputum/microbiology
Tuberculosis/diagnosis/microbiology
DNA, Bacterial
Reagent Kits, Diagnostic
Figure
Cited by 3 articles
-
Molecular Diagnosis of Tuberculosis
Fariz Nurwidya, Diah Handayani, Erlina Burhan, Faisal Yunus
Chonnam Med J. 2018;54(1):1-9. doi: 10.4068/cmj.2018.54.1.1.Comparison of AdvanSure TB/NTM PCR and COBAS TaqMan MTB PCR for Detection of
Mycobacterium tuberculosis Complex in Routine Clinical Practice
Won-Hyung Cho, Eun-Jeong Won, Hyun-Jung Choi, Seung-Jung Kee, Jong-Hee Shin, Dong-Wook Ryang, Soon-Pal Suh
Ann Lab Med. 2015;35(3):356-361. doi: 10.3343/alm.2015.35.3.356.Performance Evaluation of Anyplex plus MTB/NTM and AdvanSure TB/NTM for the Detection of
Mycobacterium tuberculosis and Nontuberculous Mycobacteria
Wonho Choe, Ehwa Kim, Seo Yeon Park, Jeong Don Chae
Ann Clin Microbiol. 2015;18(2):44-51. doi: 10.5145/ACM.2015.18.2.44.
Reference
-
1. Blakemore R, Nabeta P, Davidow AL, Vadwai V, Tahirli R, Munsamy V, et al. A multisite assessment of the quantitative capabilities of the Xpert MTB/RIF assay. Am J Respir Crit Care Med. 2011; 184:1076–1084. PMID: 21836139.
Article2. Theron G, Pinto L, Peter J, Mishra HK, Mishra HK, van Zyl-Smit R, et al. The use of an automated quantitative polymerase chain reaction (Xpert MTB/RIF) to predict the sputum smear status of tuberculosis patients. Clin Infect Dis. 2012; 54:384–388. PMID: 22139854.
Article3. Bang HI, Choi TY, Shin JW. Comparison of Ogawa Media, BACTEC MGIT 960 System and TB/NTM real-time PCR for detecting Mycobacterium species. Tuberc Respir Dis. 2011; 71:249–253.4. Hwang S, Oh KJ, Jang IH, Uh Y, Yoon KJ, Kim HY, et al. Evaluation of the diagnostic performance of the AdvanSure TB/NTM real-time PCR kit for detection of mycobacteria. Korean J Clin Microbiol. 2011; 14:55–59.
Article5. Kim YJ, Park MY, Kim SY, Cho SA, Hwang SH, Kim HH, et al. Evaluation of the performances of AdvanSure TB/NTM real time PCR kit for detection of mycobacteria in respiratory specimens. Korean J Lab Med. 2008; 28:34–38. PMID: 18309253.
Article6. Clinical and Laboratory Standards Institute. Laboratory detection and identification of mycobacteria; approved guideline, M48-A. Wayne, PA: Clinical and Laboratory Standards Institute;2008. p. 27.7. Centers for Disease Control and Prevention. AFB microscopy: characteristics, strategies and interpretation. Updated on Nov 2003. http://wwwn.cdc.gov/dls/ILA/Training%20Workshops/Kenya1103/Ch3/Module5microstrat.rtf.8. Theus S, Eisenach K, Fomukong N, Silver RF, Cave MD. Beijing family Mycobacterium tuberculosis strains differ in their intracellular growth in THP-1 macrophages. Int J Tuberc Lung Dis. 2007; 11:1087–1093. PMID: 17945065.9. Choi GE, Jang MH, Song EJ, Jeong SH, Kim JS, Lee WG, et al. IS6110-restriction fragment length polymorphism and spoligotyping analysis of Mycobacterium tuberculosis clinical isolates for infestigating epidemiologic distribution in Korea. J Korean Med Sci. 2010; 25:1716–1721. PMID: 21165284.10. Park YK, Kang HY, Lim JG, Ha JS, Cho JO, Lee KC, et al. Analysis of DNA fingerprints of Mycobacterium tuberculosis isolates from patients registered at health center in Gyeonggi Province in 2004. Tuberc Respir Dis. 2006; 60:290–296.11. Li L, Mahan CS, Palaci M, Horter L, Loeffelholz L, Johnson JL, et al. Sputum Mycobacterium tuberculosis mRNA as a marker of bacteriologic clearance in response to antituberculosis therapy. J Clin Microbiol. 2010; 48:46–51. PMID: 19923475.12. Hernández-Pando R, Jeyanathan M, Mengistu G, Aguilar D, Orozco H, Harboe M, et al. Persistence of DNA from Mycobacterium tuberculosis in superficially normal lung tissue during latent infection. Lancet. 2000; 356:2133–2138. PMID: 11191539.13. Behr MA, Warren SA, Salamon H, Hopewell PC, de Leon AP, Daley CL, et al. Transmission of Mycobacterium tuberculosis from patients smear-negative for acid-fast bacilli. Lancet. 1999; 353:444–449. PMID: 9989714.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of the AdvanSure TB/NTM Plus Real-Time PCR Assay for the Simultaneous Detection of Mycobacterium tuberculosis and Nontuberculous Mycobacteria from Clinical Specimens
- Evaluation of the Performances of AdvanSure TB/NTM Real Time PCR Kit for Detection of Mycobacteria in Respiratory Specimens
- Evaluation of the Diagnostic Performance of the AdvanSure TB/NTM Real-Time PCR Kit for Detection of Mycobacteria
- Head-to-Head Comparison between Xpert MTB/RIF Assay and Real-Time Polymerase Chain Reaction Assay Using Bronchial Washing Specimens for Tuberculosis Diagnosis
- Performance Evaluation of Anyplex plus MTB/NTM and AdvanSure TB/NTM for the Detection of Mycobacterium tuberculosis and Nontuberculous Mycobacteria