Clin Exp Vaccine Res.
2014 Jul;3(2):117-127. 10.7774/cevr.2014.3.2.117.
Evolutionary dynamics of highly pathogenic avian influenza A/H5N1 HA clades and vaccine implementation in Vietnam
- Affiliations
-
- 1Department of Immunology, Institute of Biotechnology, Vietnam Academy of Science and Technology, Hanoi, Vietnam. imibtvn@gmail.com
- KMID: 1730615
- DOI: http://doi.org/10.7774/cevr.2014.3.2.117
Abstract
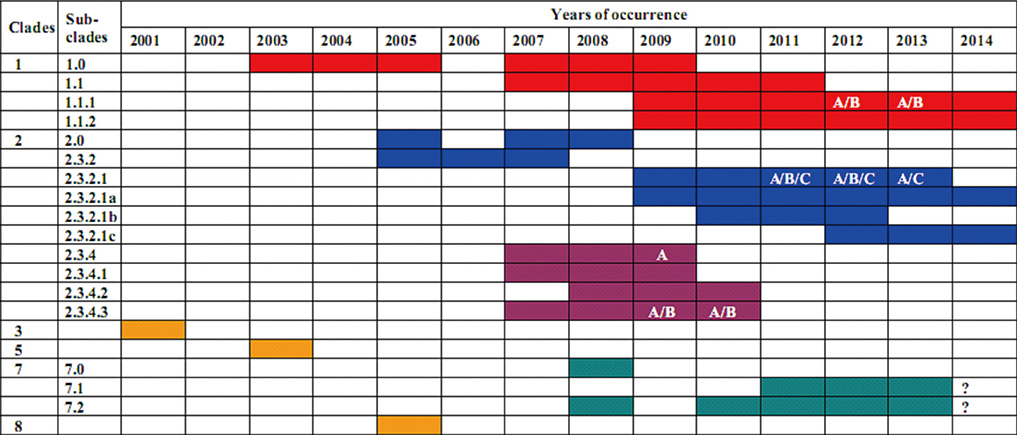
- Based on hemagglutinin (HA) and neuraminidase (NA), influenza A virus is divided into 18 different HA (H1 to H18) and 11 NA types (N1 to N11), opening the possibility for reassortment between the HA and NA genes to generate new HxNy subtypes (where x could be any HA and y is any NA, possibly). In recent four years, since 2010, highly pathogenic avian influenza (HPAI) viruses of H5N1 subtype (HPAI A/H5N1) have become highly enzootic and dynamically evolved to form multiple H5 HA clades, particularly in China, Vietnam, Indonesia, Egypt, Cambodia, and Bangladesh. So far, after more than 10 years emerged in Vietnam (since late 2003), HPAI A/H5N1 is still posing a potential risk of causing outbreaks in poultry, with high frequency of annual endemics. Intragenic variation (referred to as antigenic drift) in HA (e.g., H5) has given rise to form numerous clades, typically marking the major timelines of the evolutionary status and vaccine application in each period. The dominance of genetically and antigenically diversified clade 2.3.2.1 (of subgroups a, b, c), clade 1.1 (1.1.1/1.1.2) and re-emergence of clade 7.1/7.2 at present, has urged Vietnam to the need for dynamically applied antigenicity-matching vaccines, i.e., the plan of importing Re-6 vaccine for use in 2014, in parallel use of Re-1/Re-5 since 2006. In this review, we summarize evolutionary features of HPAI A/H5N1 viruses and clade formation during recent 10 years (2004-2014). Dynamic of vaccine implementation in Vienam is also remarked.
Keyword
MeSH Terms
Figure
Reference
-
1. Shaw M, Palese P. Orthomyxoviridae. In : Knipe DM, Howley P, editors. Field virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins;2013. Vol. 1:p. 1151–1185.2. Rabadan R, Levine AJ, Robins H. Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes. J Virol. 2006; 80:11887–11891.
Article3. Sonnberg S, Webby RJ, Webster RG. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 2013; 178:63–77.
Article4. Krumbholz A, Philipps A, Oehring H, et al. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol. 2011; 200:69–75.
Article5. Wise HM, Foeglein A, Sun J, et al. A complicated message: Identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J Virol. 2009; 83:8021–8031.
Article6. Jagger BW, Wise HM, Kash JC, et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012; 337:199–204.
Article7. Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. Identification of novel influenza A virus proteins translated from PA mRNA. J Virol. 2013; 87:2455–2462.
Article8. Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012; 109:4269–4274.
Article9. Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013; 9:e1003657.
Article10. Centers for Disease Control and Prevention. Types of influenza viruses [Internet]. Atlanta: Centers for Disease Control and Prevention;2013. cited 2013 Apr 19. Available from: http://www.cdc.gov/flu/about/viruses/types.htm.11. ElHefnawi M, Sherif FF. Accurate classification and hemagglutinin amino acid signatures for influenza A virus host-origin association and subtyping. Virology. 2014; 449:328–338.
Article12. Lu L, Lycett SJ, Leigh Brown AJ. Reassortment patterns of avian influenza virus internal segments among different subtypes. BMC Evol Biol. 2014; 14:16.
Article13. Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci U S A. 2009; 106:21312–21316.
Article14. Mair CM, Ludwig K, Herrmann A, Sieben C. Receptor binding and pH stability - how influenza A virus hemagglutinin affects host-specific virus infection. Biochim Biophys Acta. 2014; 1838:1153–1168.
Article15. Lu X, Shi Y, Gao F, et al. Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an alpha-helix structure in its cleavage site with inefficient HA1/HA2 cleavage. J Virol. 2012; 86:12861–12870.
Article16. Tharakaraman K, Raman R, Viswanathan K, et al. Structural determinants for naturally evolving H5N1 hemagglutinin to switch its receptor specificity. Cell. 2013; 153:1475–1485.
Article17. Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett. 2013; 587:1206–1214.
Article18. Hutchinson EC, Fodor E. Transport of the influenza virus genome from nucleus to nucleus. Viruses. 2013; 5:2424–2446.
Article19. Gao Q, Chou YY, Doganay S, Vafabakhsh R, Ha T, Palese P. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J Virol. 2012; 86:7043–7051.
Article20. Wang C, Zhang Y, Wu B, et al. Evolutionary characterization of the pandemic H1N1/2009 influenza virus in humans based on non-structural genes. PLoS One. 2013; 8:e56201.
Article21. Lei F, Shi W. Prospective of genomics in revealing transmission, reassortment and evolution of wildlife-borne Avian Influenza A (H5N1) viruses. Curr Genomics. 2011; 12:466–474.
Article22. Runstadler J, Hill N, Hussein IT, Puryear W, Keogh M. Connecting the study of wild influenza with the potential for pandemic disease. Infect Genet Evol. 2013; 17:162–187.
Article23. To KK, Tsang AK, Chan JF, Cheng VC, Chen H, Yuen KY. Emergence in China of human disease due to avian influenza A(H10N8): cause for concern? J Infect. 2014; 68:205–215.
Article24. Baz M, Luke CJ, Cheng X, Jin H, Subbarao K. H5N1 vaccines in humans. Virus Res. 2013; 178:78–98.
Article25. Spackman E, Swayne DE. Vaccination of gallinaceous poultry for H5N1 highly pathogenic avian influenza: current questions and new technology. Virus Res. 2013; 178:121–132.
Article26. Li C, Bu Z, Chen H. Avian influenza vaccines against H5N1 'bird flu'. Trends Biotechnol. 2014; 32:147–156.
Article27. Subbarao K, Katz JM. Influenza vaccines generated by reverse genetics. Curr Top Microbiol Immunol. 2004; 283:313–342.
Article28. Kreibich A, Stech J, Mettenleiter TC, Stech O. Simultaneous one-tube full-length amplification of the NA, NP, M, and NS genes of influenza A viruses for reverse genetics. J Virol Methods. 2009; 159:308–310.
Article29. Krammer F, Cox RJ. The emergence of H7N9 viruses: a chance to redefine correlates of protection for influenza virus vaccines. Expert Rev Vaccines. 2013; 12:1369–1372.
Article30. World Health Organization. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness [Internet]. Geneva: World Health Organization;2014. cited 2014 Feb 20. Available from: http://www.who.int/influenza/vaccines/virus/201402_h5h7h9h10_vaccinevirusupdate.pdf?ua=1.31. Velkov T, Ong C, Baker MA, et al. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol. 2013; 56:705–719.
Article32. Imai M, Kawaoka Y. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol. 2012; 2:160–167.
Article33. Galloway SE, Reed ML, Russell CJ, Steinhauer DA. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog. 2013; 9:e1003151.
Article34. Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013; 178:99–113.
Article35. Li KS, Guan Y, Wang J, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004; 430:209–213.
Article36. Smith GJ, Naipospos TS, Nguyen TD, et al. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology. 2006; 350:258–268.
Article37. Macken CA, Webby RJ, Bruno WJ. Genotype turnover by reassortment of replication complex genes from avian influenza A virus. J Gen Virol. 2006; 87(Pt 10):2803–2815.
Article38. Wu WL, Chen Y, Wang P, et al. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. J Virol. 2008; 82:1798–1807.
Article39. Duan L, Bahl J, Smith GJ, et al. The development and genetic diversity of H5N1 influenza virus in China, 1996-2006. Virology. 2008; 380:243–254.
Article40. Wan XF, Nguyen T, Davis CT, et al. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One. 2008; 3:e3462.
Article41. Nguyen TD, Nguyen TV, Vijaykrishna D, et al. Multiple sublineages of influenza A virus (H5N1), Vietnam, 2005-2007. Emerg Infect Dis. 2008; 14:632–636.
Article42. Nguyen T, Rivailler P, Davis CT, et al. Evolution of highly pathogenic avian influenza (H5N1) virus populations in Vietnam between 2007 and 2010. Virology. 2012; 432:405–416.
Article43. Creanga A, Thi Nguyen D, Gerloff N, et al. Emergence of multiple clade 2.3.2.1 influenza A (H5N1) virus subgroups in Vietnam and detection of novel reassortants. Virology. 2013; 444:12–20.
Article44. World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization (WHO/OIE/FAO) H5N1 Evolution Working Group. Revised and updated nomenclature for highly pathogenic avian influenza A (H5N1) viruses. Influenza Other Respir Viruses. 2014; 8:384–388.45. Inui K. Epi-zone approach for HPAI H5N1 surveillance and control [Internet]. cited 2014 May 1. Available from: http://www.rr-asia.oie.int/fileadmin/Regional_Representation/Programme/H_HPAI/JTF/2012_5th_ExpMeeting/2-1_Dr_Inui.pdf.46. Long NT, Thanh TT, van Doorn HR, et al. Recent avian influenza virus A/H5N1 evolution in vaccinated and unvaccinated poultry from farms in Southern Vietnam, January-March 2010. Transbound Emerg Dis. 2011; 58:537–543.
Article47. Carrel MA, Emch M, Nguyen T, Todd Jobe R, Wan XF. Population-environment drivers of H5N1 avian influenza molecular change in Vietnam. Health Place. 2012; 18:1122–1131.
Article48. Cha RM, Smith D, Shepherd E, et al. Suboptimal protection against H5N1 highly pathogenic avian influenza viruses from Vietnam in ducks vaccinated with commercial poultry vaccines. Vaccine. 2013; 31:4953–4960.
Article49. Gu M, Zhao G, Zhao K, et al. Novel variants of clade 2.3.4 highly pathogenic avian influenza A(H5N1) viruses, China. Emerg Infect Dis. 2013; 19:2021–2024.
Article50. Kang HM, Batchuluun D, Kim MC, et al. Genetic analyses of H5N1 avian influenza virus in Mongolia, 2009 and its relationship with those of eastern Asia. Vet Microbiol. 2011; 147:170–175.
Article51. Hu X, Liu D, Wang M, et al. Clade 2.3.2 avian influenza virus (H5N1), Qinghai Lake region, China, 2009-2010. Emerg Infect Dis. 2011; 17:560–562.
Article52. Gilbert M, Jambal L, Karesh WB, et al. Highly pathogenic avian influenza virus among wild birds in Mongolia. PLoS One. 2012; 7:e44097.
Article53. Uchida Y, Suzuki Y, Shirakura M, et al. Genetics and infectivity of H5N1 highly pathogenic avian influenza viruses isolated from chickens and wild birds in Japan during 2010-11. Virus Res. 2012; 170:109–117.
Article54. Nagarajan S, Tosh C, Smith DK, et al. Avian influenza (H5N1) virus of clade 2.3.2 in domestic poultry in India. PLoS One. 2012; 7:e31844.
Article55. Islam MR, Haque ME, Giasuddin M, et al. New introduction of clade 2.3.2.1 avian influenza virus (H5N1) into Bangladesh. Transbound Emerg Dis. 2012; 59:460–463.
Article56. Marinova-Petkova A, Georgiev G, Seiler P, et al. Spread of influenza virus A (H5N1) clade 2.3.2.1 to Bulgaria in common buzzards. Emerg Infect Dis. 2012; 18:1596–1602.
Article57. Dharmayanti NL, Hartawan R, Pudjiatmoko , et al. Genetic characterization of clade 2.3.2.1 avian influenza A(H5N1) viruses, Indonesia, 2.0.1.2. Emerg Infect Dis. 2014; 20:671–674.
Article58. Xu L, Bao L, Yuan J, et al. Antigenicity and transmissibility of a novel clade 2.3.2.1 avian influenza H5N1 virus. J Gen Virol. 2013; 94(Pt 12):2616–2626.
Article59. Nguyen T, Davis CT, Stembridge W, et al. Characterization of a highly pathogenic avian influenza H5N1 virus sublineage in poultry seized at ports of entry into Vietnam. Virology. 2009; 387:250–256.
Article60. Le TH, Le KX, Cuong PV, et al. Adjuvant effects of Sophy β-glucan on H5N1 and H5N2 vaccination using a mouse model. Trop Med Health. 2010; 38:23–27.
Article61. Le T, Le T, Doan TH, et al. The adjuvant effect of Sophy beta-glucan to the antibody response in poultry immunized by the avian influenza A H5N1 and H5N2 vaccines. J Microbiol Biotechnol. 2011; 21:405–411.
Article62. Davis CT, Balish AL, O'Neill E, et al. Detection and characterization of clade 7 high pathogenicity avian influenza H5N1 viruses in chickens seized at ports of entry and live poultry markets in Vietnam. Avian Dis. 2010; 54:1 Suppl. 307–312.
Article63. Tung DH, Van Quyen D, Nguyen T, Xuan HT, Nam TN, Duy KD. Molecular characterization of a H5N1 highly pathogenic influenza virus clade 2.3.2.1b circulating in Vietnam in 2011. Vet Microbiol. 2013; 165:341–348.
Article64. Bui VN, Dao TD, Nguyen TT, et al. Pathogenicity of an H5N1 avian influenza virus isolated in Vietnam in 2012 and reliability of conjunctival samples for diagnosis of infection. Virus Res. 2014; 179:125–132.
Article65. Tien NN. Current situation of avian influenza, period of 2009-2012, and actual prevention solutions. Vet Sci Tech. 2013; 20:82–90.66. Hanh TX, Thi P, Thuy DH, et al. Testing the effect of the NAVET-Vifluvac vaccine against A/H5N1 viruses of clade 1.1 and 2.3.2.1c by challenging and direct contact of virulent strains. Vet Sci Tech. 2013; 20:22–29.67. Li Y, Liu L, Zhang Y, et al. New avian influenza virus (H5N1) in wild birds, Qinghai, China. Emerg Infect Dis. 2011; 17:265–267.
Article68. Nga NT, Le TH. Clarification of new clades 1.1 and 2.3.2.1 of avian influenza virus (H5N1) in Vietnam based on genetic and phylogenetic analysis of hemagglutinin (H5) genes of strains collected during 2004-2011. Vet Sci Tech. 2012; 19:20–28.69. Nga NT, Khien DV, Le TH. Confirmation of clades 2.3.2.1a and 2.3.2.1c of A/H5N1 viruses isolated in 2013 in Vietnam. Vet Sci Tech. 2013; 20:16–21.70. Gerloff NA, Khan SU, Balish A, et al. Multiple reassortment events among highly pathogenic avian influenza A(H5N1) viruses detected in Bangladesh. Virology. 2014; 450-451:297–307.
Article71. Phuong do Q, Dung NT, Jorgensen PH, Handberg KJ, Vinh NT, Christensen JP. Susceptibility of Muscovy (Cairina Moschata) and mallard ducks (Anas Platyrhynchos) to experimental infections by different genotypes of H5N1 avian influenza viruses. Vet Microbiol. 2011; 148:168–174.
Article72. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 60. Mol Biol Evol. 2013; 30:2725–2729.
Article73. Sorn S, Sok T, Ly S, et al. Dynamic of H5N1 virus in Cambodia and emergence of a novel endemic sub-clade. Infect Genet Evol. 2013; 15:87–94.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Avian Influenza: Should China Be Alarmed?
- The Current Trend of Avian Influenza Viruses in Bioinformatics Research
- Recent outbreaks of highly pathogenic avian influenza viruses in South Korea
- Control of Avian Influenza: Calls for International Collaboration
- H5 and H9 subtypes of Avian Influenza Viruses are Real Threat To Humans