J Korean Med Sci.
2024 Apr;39(15):e136. 10.3346/jkms.2024.39.e136.
Characterization of Ceftriaxone-Resistant Haemophilus influenzae Among Korean Children
- Affiliations
-
- 1Department of Pediatrics, Asan Medical Center Children’s Hospital, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2555469
- DOI: http://doi.org/10.3346/jkms.2024.39.e136
Abstract
- Background
Haemophilus influenzae is a frequently encountered pathogen responsible for respiratory tract infections in children. Following the detection of ceftriaxone-resistant H. influenzae at our institution, we aimed to investigate the resistance mechanisms of ceftriaxone in H. influenzae, with a particular focus on alterations in penicillin-binding protein 3 (PBP3) and β-lactamase production.
Methods
Among H. influenzae isolates collected at Asan Medical Center Children’s Hospital from March 2014 to April 2019, ceftriaxone-resistant strains by the disk-diffusion test were included. Ceftriaxone minimum inhibitory concentrations (MICs) were determined using the E-test according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. The presence of β-lactamase was assessed through cefinase test and TEM-1/ROB-1 polymerase chain reaction (PCR). PBP3 alterations were explored via ftsI gene sequencing.
Results
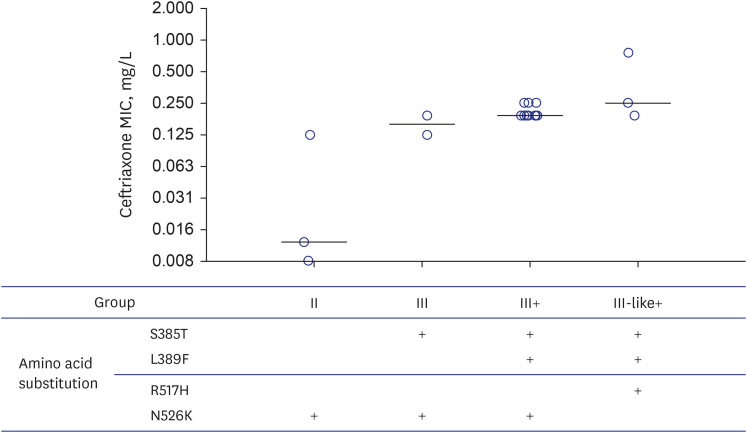
Out of the 68 collected strains, 21 exhibited resistance to ceftriaxone in disk diffusion tests. Two strains were excluded due to failed subculture. Among 19 ceftriaxoneresistant H. influenzae isolates, eighteen were non-typeable H. influenzae, and twelve were positive for TEM-1 PCR. Isolates were classified into groups II (harboring only N526K, n = 3), III (N526K+S385T, n = 2), III+ (S385T+L389F+N526K, n = 11), and III-like+ (S385T+L389F+R517H, n = 3) according to the PBP3 alteration pattern. With a median ceftriaxone MIC of 0.190 mg/L (range, 0.008–0.750), the median ceftriaxone MIC was the highest in group III-like+ (0.250 mg/L), followed by groups III+ (0.190 mg/L), III (0.158 mg/L), and II (0.012 mg/L). All three strains belonging to group II, which did not harbor the S385T substitution, had ceftriaxone MICs of ≤ 0.125 mg/L.
Conclusion
The emergence of ceftriaxone-resistant H. influenzae with ceftriaxone MIC values of up to 0.75 mg/L was observed even in children in South Korea, with most associated with S385T and L389F substitutions. The N526K mutation alone does not significantly impact ceftriaxone resistance. Further large-scale studies are essential to investigate changes in antibiotic resistance patterns and factors influencing antibiotic resistance in H. influenzae isolated from pediatric patients in Korea.
Figure
Reference
-
1. Tristram S, Jacobs MR, Appelbaum PC. Antimicrobial resistance in Haemophilus influenzae . Clin Microbiol Rev. 2007; 20(2):368–389. PMID: 17428889.2. Langereis JD, de Jonge MI. Invasive disease caused by Nontypeable Haemophilus influenzae . Emerg Infect Dis. 2015; 21(10):1711–1718. PMID: 26407156.3. Kim JS, Jang YT, Kim JD, Park TH, Park JM, Kilgore PE, et al. Incidence of Haemophilus influenzae type b and other invasive diseases in South Korean children. Vaccine. 2004; 22(29-30):3952–3962. PMID: 15364444.4. Wang S, Tafalla M, Hanssens L, Dolhain J. A review of Haemophilus influenzae disease in Europe from 2000-2014: challenges, successes and the contribution of hexavalent combination vaccines. Expert Rev Vaccines. 2017; 16(11):1095–1105. PMID: 28971707.5. Gilsdorf JR. Hib vaccines: their impact on Haemophilus influenzae Type b disease. J Infect Dis. 2021; 224(12):Suppl 2. S321–S330. PMID: 34590133.6. Kim IS, Ki CS, Kim S, Oh WS, Peck KR, Song JH, et al. Diversity of ampicillin resistance genes and antimicrobial susceptibility patterns in Haemophilus influenzae strains isolated in Korea. Antimicrob Agents Chemother. 2007; 51(2):453–460. PMID: 17116681.7. Hoban D, Felmingham D. The PROTEKT surveillance study: antimicrobial susceptibility of Haemophilus influenzae and Moraxella catarrhalis from community-acquired respiratory tract infections. J Antimicrob Chemother. 2002; 50(Suppl S1):49–59. PMID: 12239228.8. Bae S, Lee J, Lee J, Kim E, Lee S, Yu J, et al. Antimicrobial resistance in Haemophilus influenzae respiratory tract isolates in Korea: results of a nationwide acute respiratory infections surveillance. Antimicrob Agents Chemother. 2010; 54(1):65–71. PMID: 19884366.9. Mason EO Jr, Kaplan SL, Lamberth LB, Hinds DB, Kvernland SJ, Loiselle EM, et al. Serotype and ampicillin susceptibility of Haemophilus influenzae causing systemic infections in children: 3 years of experience. J Clin Microbiol. 1982; 15(4):543–546. PMID: 6978349.10. Skaare D, Anthonisen IL, Kahlmeter G, Matuschek E, Natås OB, Steinbakk M, et al. Emergence of clonally related multidrug resistant Haemophilus influenzae with penicillin-binding protein 3-mediated resistance to extended-spectrum cephalosporins, Norway, 2006 to 2013. Euro Surveill. 2014; 19(49):20986. PMID: 25523969.11. Skaare D, Allum AG, Anthonisen IL, Jenkins A, Lia A, Strand L, et al. Mutant ftsI genes in the emergence of penicillin-binding protein-mediated beta-lactam resistance in Haemophilus influenzae in Norway. Clin Microbiol Infect. 2010; 16(8):1117–1124. PMID: 19737286.12. Park C, Kim KH, Shin NY, Byun JH, Kwon EY, Lee JW, et al. Genetic diversity of the ftsI gene in β-lactamase-nonproducing ampicillin-resistant and β-lactamase-producing amoxicillin-/clavulanic acid-resistant nasopharyngeal Haemophilus influenzae strains isolated from children in South Korea. Microb Drug Resist. 2013; 19(3):224–230. PMID: 23308379.13. Kubota T, Higa F, Kusano N, Nakasone I, Haranage S, Tateyama M, et al. Genetic analyses of beta-lactamase negative ampicillin-resistant strains of Haemophilus influenzae isolated in Okinawa, Japan. Jpn J Infect Dis. 2006; 59(1):36–41. PMID: 16495632.14. Osaki Y, Sanbongi Y, Ishikawa M, Kataoka H, Suzuki T, Maeda K, et al. Genetic approach to study the relationship between penicillin-binding protein 3 mutations and Haemophilus influenzae beta-lactam resistance by using site-directed mutagenesis and gene recombinants. Antimicrob Agents Chemother. 2005; 49(7):2834–2839. PMID: 15980357.15. Hasegawa K, Chiba N, Kobayashi R, Murayama SY, Iwata S, Sunakawa K, et al. Rapidly increasing prevalence of beta-lactamase-nonproducing, ampicillin-resistant Haemophilus influenzae type b in patients with meningitis. Antimicrob Agents Chemother. 2004; 48(5):1509–1514. PMID: 15105098.16. Lee E, Park S, Kim M, Lee J. Trend of antibiotic susceptibility of Haemophilus influenzae isolated from children, 2014–2019. Pediatr Infect Vaccine. 2020; 27(3):147–157.17. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 14.0, 2024. Updated 2024. Accessed March 18, 2024. http://www.eucast.org .18. Clifford RJ, Milillo M, Prestwood J, Quintero R, Zurawski DV, Kwak YI, et al. Detection of bacterial 16S rRNA and identification of four clinically important bacteria by real-time PCR. PLoS One. 2012; 7(11):e48558. PMID: 23139793.19. van Ketel RJ, de Wever B, van Alphen L. Detection of Haemophilus influenzae in cerebrospinal fluids by polymerase chain reaction DNA amplification. J Med Microbiol. 1990; 33(4):271–276. PMID: 2258914.20. Han MS, Jung HJ, Lee HJ, Choi EH. Increasing prevalence of group III penicillin-binding protein 3 mutations conferring high-level resistance to beta-lactams among nontypeable Haemophilus influenzae isolates from children in Korea. Microb Drug Resist. 2019; 25(4):567–576. PMID: 30484742.21. Ubukata K, Shibasaki Y, Yamamoto K, Chiba N, Hasegawa K, Takeuchi Y, et al. Association of amino acid substitutions in penicillin-binding protein 3 with beta-lactam resistance in beta-lactamase-negative ampicillin-resistant Haemophilus influenzae . Antimicrob Agents Chemother. 2001; 45(6):1693–1699. PMID: 11353613.22. García-Cobos S, Campos J, Lázaro E, Román F, Cercenado E, García-Rey C, et al. Ampicillin-resistant non-beta-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob Agents Chemother. 2007; 51(7):2564–2573. PMID: 17470649.23. Kwak YH, Jung HS, Park SE, Park JY, Kim EC, Lee HJ, et al. Serotypes and antimicrobial susceptibility in clinical isolates of Haemophilus influenzae from Korean children in prevaccination era. J Korean Med Sci. 2000; 15(6):616–622. PMID: 11194185.24. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 34th ed. Wayne, PA, USA: Clinical and Laboratory Standards Institute;2024.25. Ferjani S, Sassi I, Saidani M, Mhiri E, Ghariani A, Boutiba Ben Boubaker I, et al. Polymorphism of ftsI gene in Haemophilus influenzae and emergence of cefotaxime resistance in two Tunisian hospitals. New Microbes New Infect. 2020; 36:100690. PMID: 32489667.26. Mizoguchi A, Hitomi S. Cefotaxime-non-susceptibility of Haemophilus influenzae induced by additional amino acid substitutions of G555E and Y557H in altered penicillin-binding protein 3. J Infect Chemother. 2019; 25(7):509–513. PMID: 30879978.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Trend of Antibiotic Susceptibility of Haemophilus influenzae Isolated from Children, 2014–2019
- A Case of Neurologic Sequelae and a Case of Peripheral Gangrene of Extremities Associated with Haemophilus influenzae Type b Meningitis
- Haemophilus influenzae vaccine
- Serotypes and antimicrobial susceptibility in clinical isolates of Haemophilus influenzae from Korean children in prevaccination era
- Natural antibody against haemophilus influenzae type b in a sample population of Korean children