Healthc Inform Res.
2024 Apr;30(2):140-146. 10.4258/hir.2024.30.2.140.
Development and Validation of Adaptable Skin Cancer Classification System Using Dynamically Expandable Representation
- Affiliations
-
- 1Department of Biomedical Systems Informatics, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2555213
- DOI: http://doi.org/10.4258/hir.2024.30.2.140
Abstract
Objectives
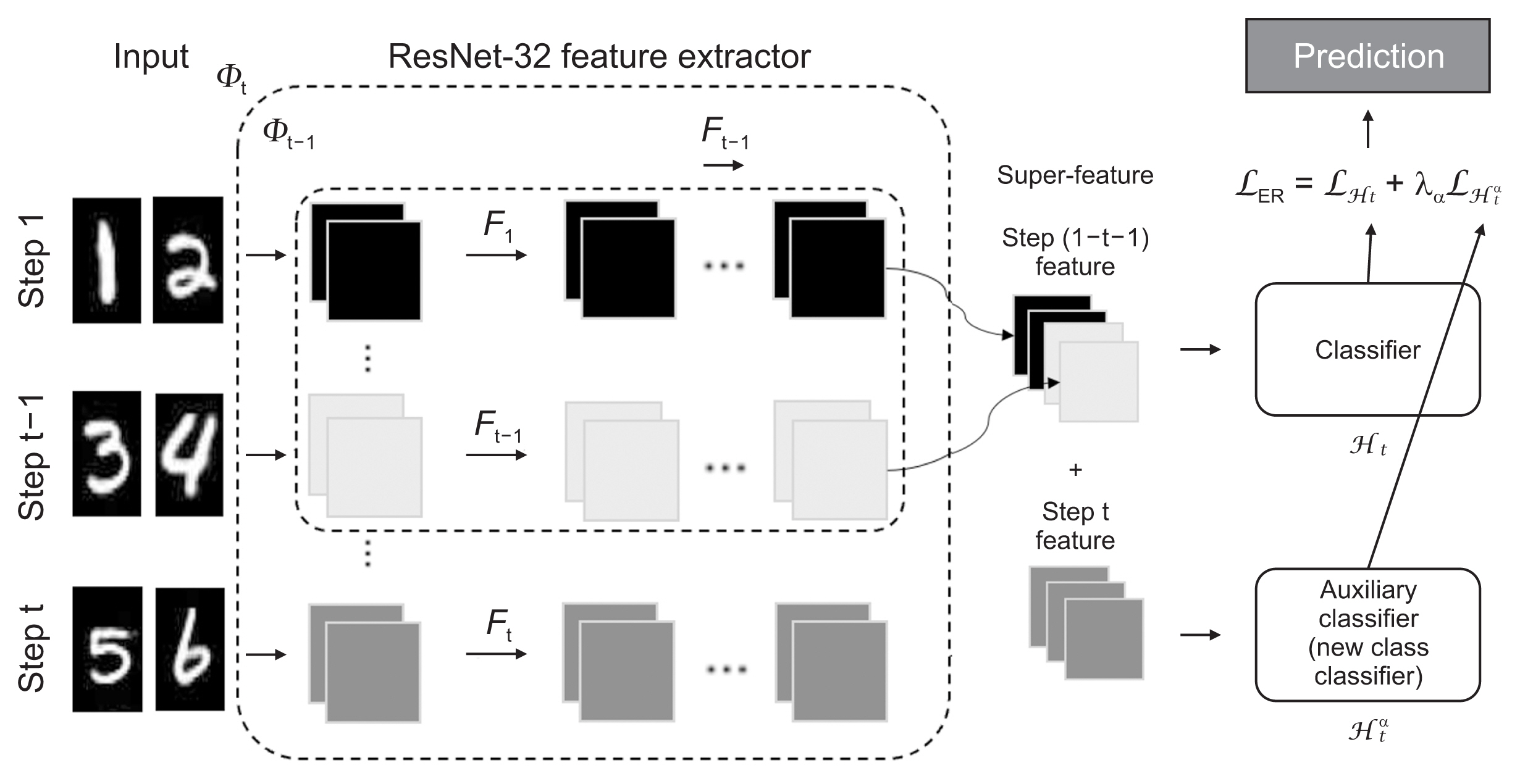
Skin cancer is a prevalent type of malignancy, necessitating efficient diagnostic tools. This study aimed to develop an automated skin lesion classification model using the dynamically expandable representation (DER) incremental learning algorithm. This algorithm adapts to new data and expands its classification capabilities, with the goal of creating a scalable and efficient system for diagnosing skin cancer.
Methods
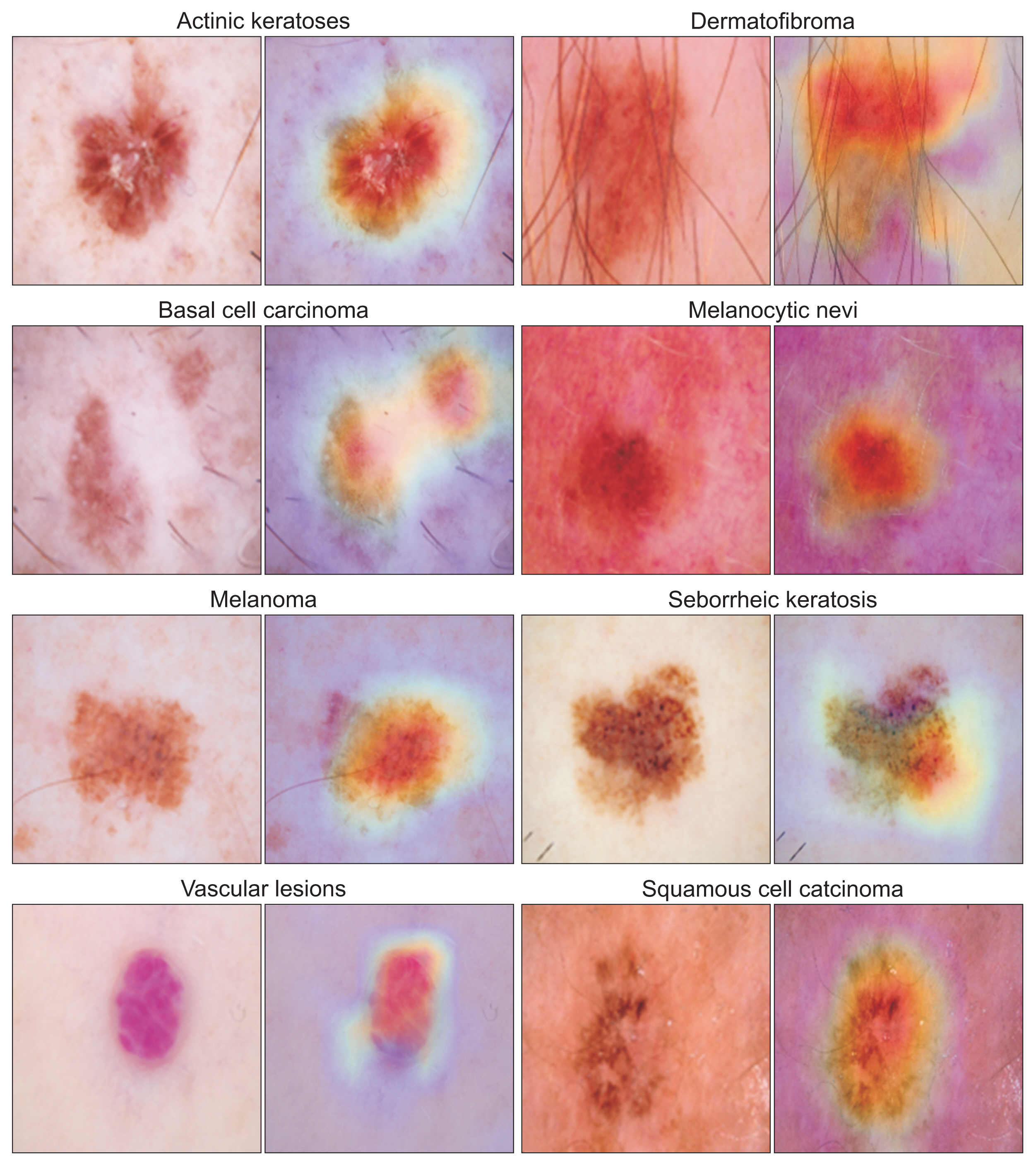
The DER model with incremental learning was applied to the HAM10000 and ISIC 2019 datasets. Validation involved two steps: initially, training and evaluating the HAM10000 dataset against a fixed ResNet-50; subsequently, performing external validation of the trained model using the ISIC 2019 dataset. The model’s performance was assessed using precision, recall, the F1-score, and area under the precision-recall curve.
Results
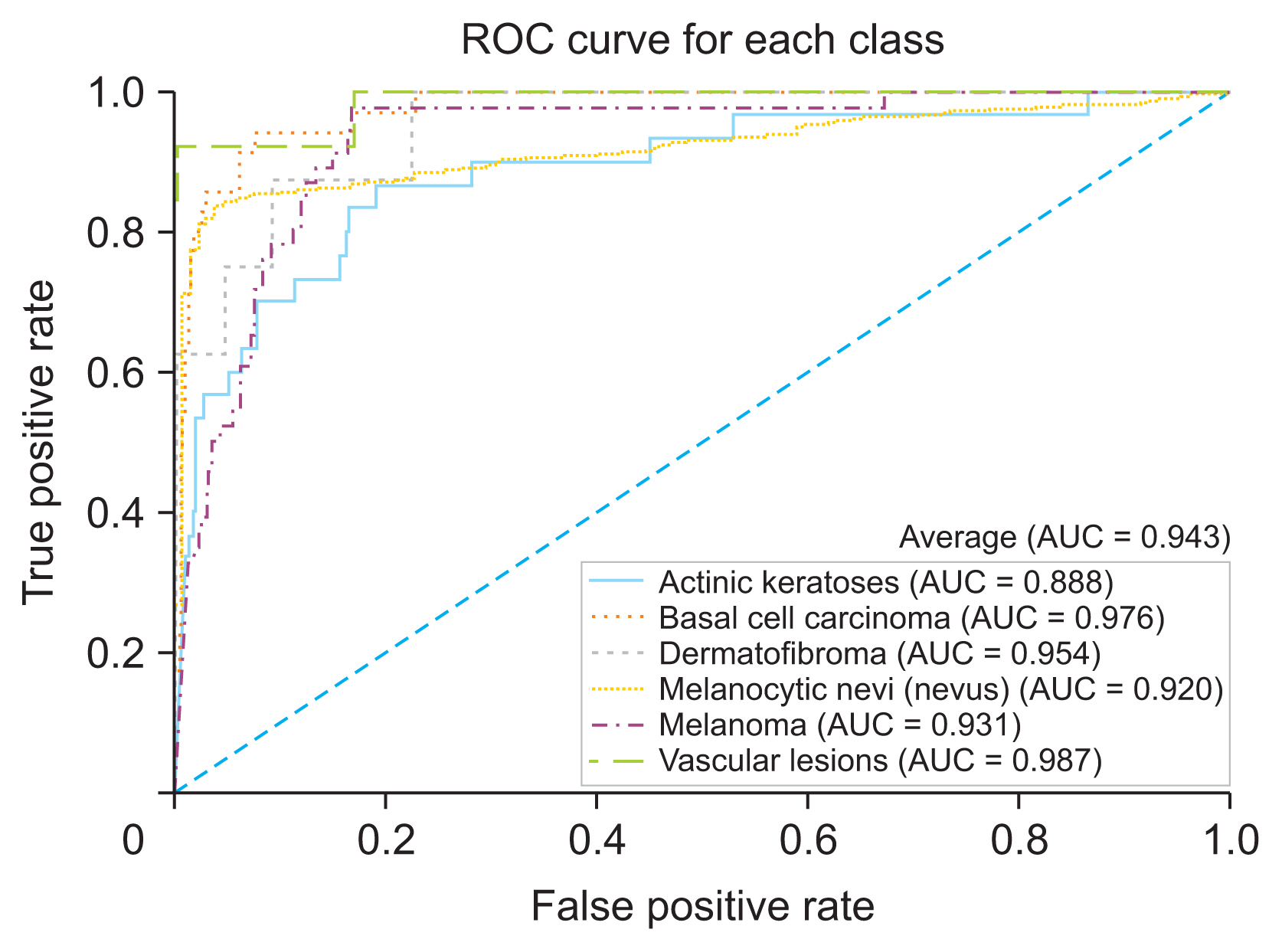
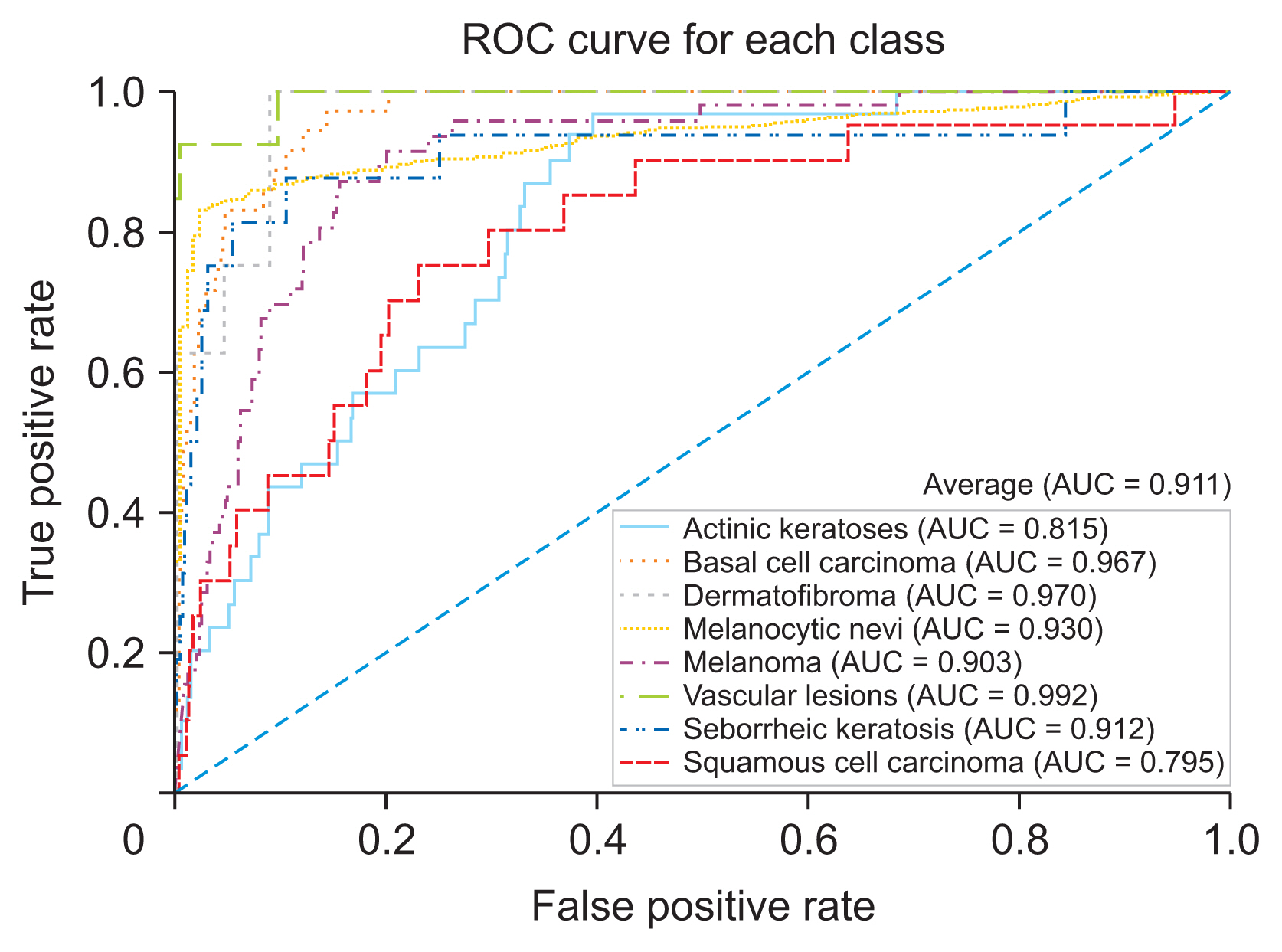
The developed skin lesion classification model demonstrated high accuracy and reliability across various types of skin lesions, achieving a weighted-average precision, recall, and F1-score of 0.918, 0.808, and 0.847, respectively. The model’s discrimination performance was reflected in an average area under the curve (AUC) value of 0.943. Further external validation with the ISIC 2019 dataset confirmed the model’s effectiveness, as shown by an AUC of 0.911.
Conclusions
This study presents an optimized skin lesion classification model based on the DER algorithm, which shows high performance in disease classification with the potential to expand its classification range. The model demonstrated robust results in external validation, indicating its adaptability to new disease classes.
Keyword
Figure
Reference
-
References
1. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017; 542(7639):115–8. https://doi.org/10.1038/nature21056.
Article2. Abedini M, Codella NC, Connell JH, Garnavi R, Merler M, Pankanti S, et al. A generalized framework for medical image classification and recognition. IBM J Res Dev. 2015; 59(2/3):1–18. https://doi.org/10.1147/JRD.2015.2390017.
Article3. Haenssle HA, Fink C, Schneiderbauer R, Toberer F, Buhl T, Blum A, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018; 29(8):1836–42. https://doi.org/10.1093/annonc/mdy166.
Article4. Kumar P, Srivastava MM. Example mining for incremental learning in medical imaging. In : Proceedings of 2018 IEEE Symposium Series on Computational Intelligence (SSCI); 2018 Nov 18–21; Bangalore, India. 48–51. https://doi.org/10.1109/SSCI.2018.8628895.
Article5. Yan S, Xie J, He X. DER: dynamically expandable representation for class incremental learning. In : Proceedings of 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jan 20–25; Nashville, TN, USA. p. 3014–23. https://doi.org/10.1109/CVPR46437.2021.00303.
Article6. Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018; 5:180161. https://doi.org/10.1038/sdata.2018.161.
Article7. Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, et al. Skin lesion analysis toward melanoma detection: a challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), hosted by the International Skin Imaging Collaboration (ISIC). In : Proceedings of 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI); 2018 Apr 4–7; Washington, DC, USA. p. 168–72. https://doi.org/10.1109/ISBI.2018.8363547.
Article8. Cubuk ED, Zoph B, Mane D, Vasudevan V, Le QV. AutoAugment: learning augmentation strategies from data. In : Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15–20; Long Beach, CA, USA. p. 113–23. https://doi.org/10.1109/CVPR.2019.00020.
Article9. Barata C, Celebi ME, Marques JS. Improving dermoscopy image classification using color constancy. IEEE J Biomed Health Inform. 2015; 19(3):1146–52. https://doi.org/10.1109/JBHI.2014.2336473.
Article10. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In : Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27 30; Las Vegas, NV, USA. p. 770–8. https://doi.org/10.1109/CVPR.2016.90.
Article11. Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013; 4(2):627–35.12. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In : Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, Italy. p. 618–26. https://doi.org/10.1109/ICCV.2017.74.
Article13. Popescu D, El-Khatib M, Ichim L. Skin lesion classification using collective intelligence of multiple neural networks. Sensors (Basel). 2022; 22(12):4399. https://doi.org/10.3390/s22124399.
Article14. Hasselmo ME. Avoiding catastrophic forgetting. Trends Cogn Sci. 2017; 21(6):407–8. https://doi.org/10.1016/j.tics.2017.04.001.
Article15. Kirkpatrick J, Pascanu R, Rabinowitz N, Veness J, Desjardins G, Rusu AA, et al. Overcoming catastrophic forgetting in neural networks. Proc Natl Acad Sci U S A. 2017; 114(13):3521–6. https://doi.org/10.1073/pnas.1611835114.
Article16. Althnian A, AlSaeed D, Al-Baity H, Samha A, Dris AB, Alzakari N, et al. Impact of dataset size on classification performance: an empirical evaluation in the medical domain. Appl Sci. 2021; 11(2):796. https://doi.org/10.3390/app11020796.
Article17. Vabalas A, Gowen E, Poliakoff E, Casson AJ. Machine learning algorithm validation with a limited sample size. PLoS One. 2019; 14(11):e0224365. https://doi.org/10.1371/journal.pone.0224365.
Article18. Lupton E. Skin: surface, substance, and design. New York (NY): Princeton Architectural Press;2007.19. Chen X, Wen J, Wu W, Peng Q, Cui X, He L. A review of factors influencing sensitive skin: an emphasis on built environment characteristics. Front Public Health. 2023; 11:1269314. https://doi.org/10.3389/fpubh.2023.1269314.
Article20. Celebi Sozener Z, Ozdel Ozturk B, Cerci P, Turk M, Gorgulu Akin B, Akdis M, et al. Epithelial barrier hypothesis: effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy. 2022; 77(5):1418–49. https://doi.org/10.1111/all.15240.
Article21. Popescu D, El-Khatib M, El-Khatib H, Ichim L. New trends in melanoma detection using neural networks: a systematic review. Sensors (Basel). 2022; 22(2):496. https://doi.org/10.3390/s22020496.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development and Validation of a Prediction Model: Application to Digestive Cancer Research
- Kyoto Classification of Gastritis and Gastric Cancer
- Anatomy and Classification of Anorectal Malformation
- Illness Representation for Pathological Gambling
- Letter to the Editor Regarding “Validation of Ultrasound and Computed Tomography-Based Risk Stratification System and Biopsy Criteria for Cervical Lymph Nodes in Preoperative Patients With Thyroid Cancer” and “Validation of CT-Based Risk Stratification System for Lymph Node Metastasis in Patients With Thyroid Cancer”