Clin Exp Otorhinolaryngol.
2024 Feb;17(1):85-97. 10.21053/ceo.2023.00026.
Development and Validation of a Pathomics Model Using Machine Learning to Predict CXCL8 Expression and Prognosis in Head and Neck Cancer
- Affiliations
-
- 1Department of Otolaryngology-Head and Neck Surgery, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai, China
- 2Department of Nursing, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai, China
- KMID: 2553059
- DOI: http://doi.org/10.21053/ceo.2023.00026
Abstract
Objectives
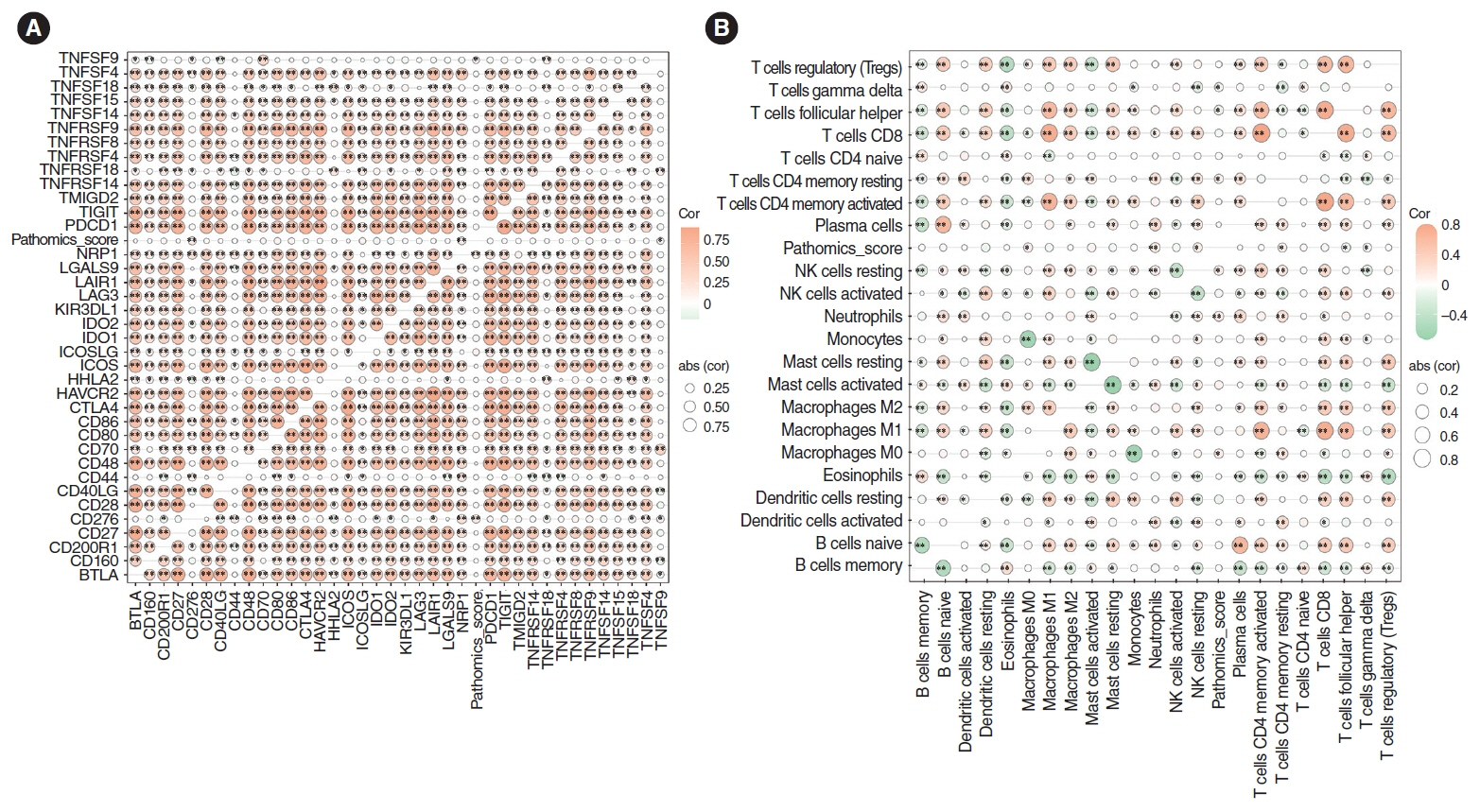
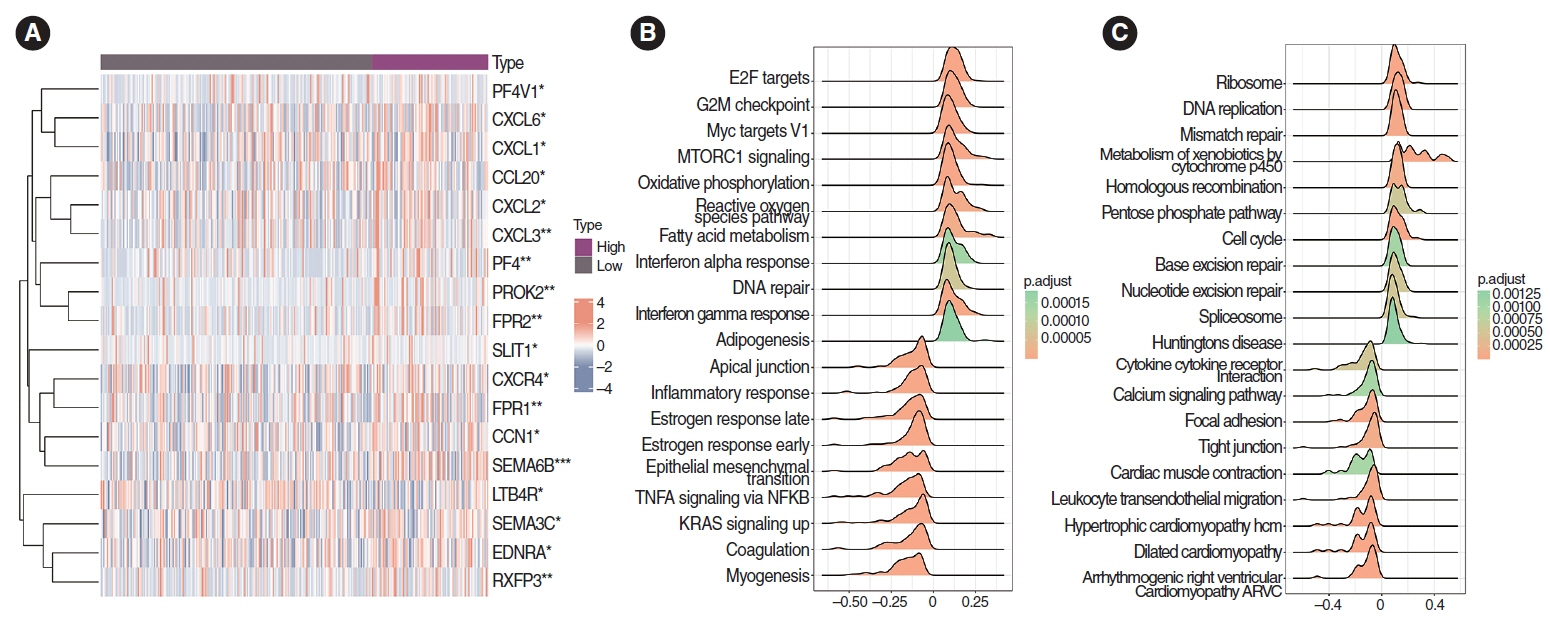
. The necessity to develop a method for prognostication and to identify novel biomarkers for personalized medicine in patients with head and neck squamous cell carcinoma (HNSCC) cannot be overstated. Recently, pathomics, which relies on quantitative analysis of medical imaging, has come to the forefront. CXCL8, an essential inflammatory cytokine, has been shown to correlate with overall survival (OS). This study examined the relationship between CXCL8 mRNA expression and pathomics features and aimed to explore the biological underpinnings of CXCL8.
Methods
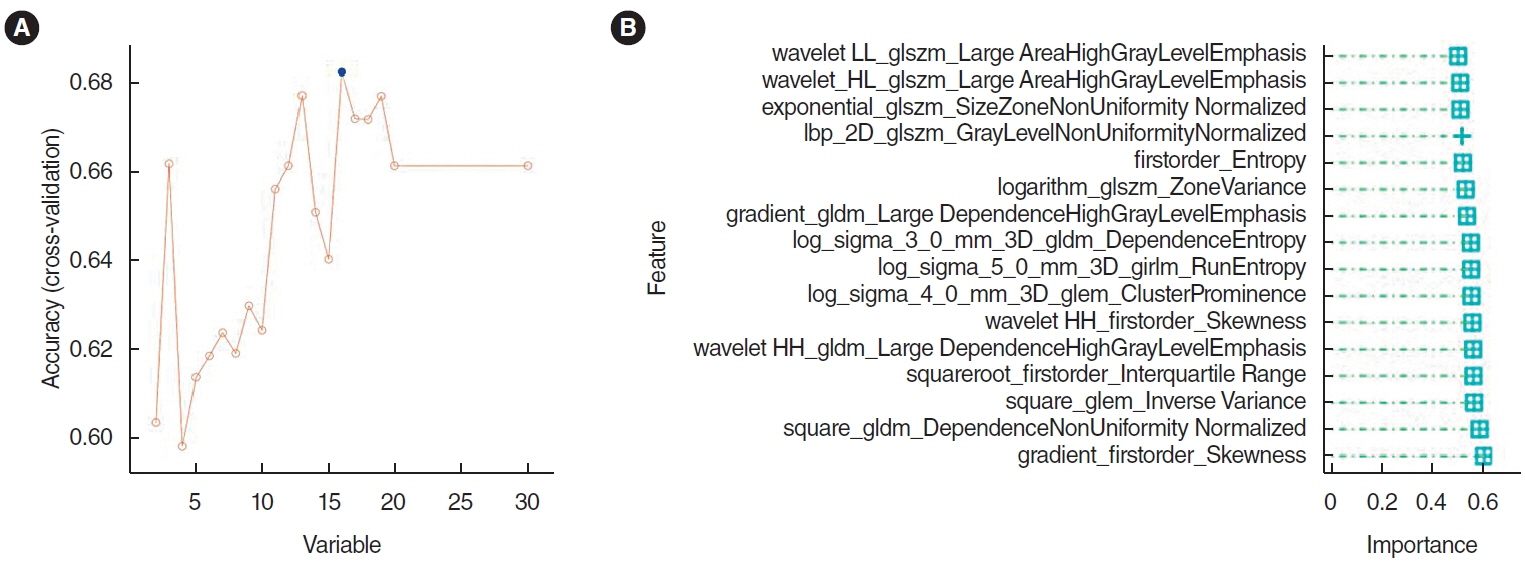
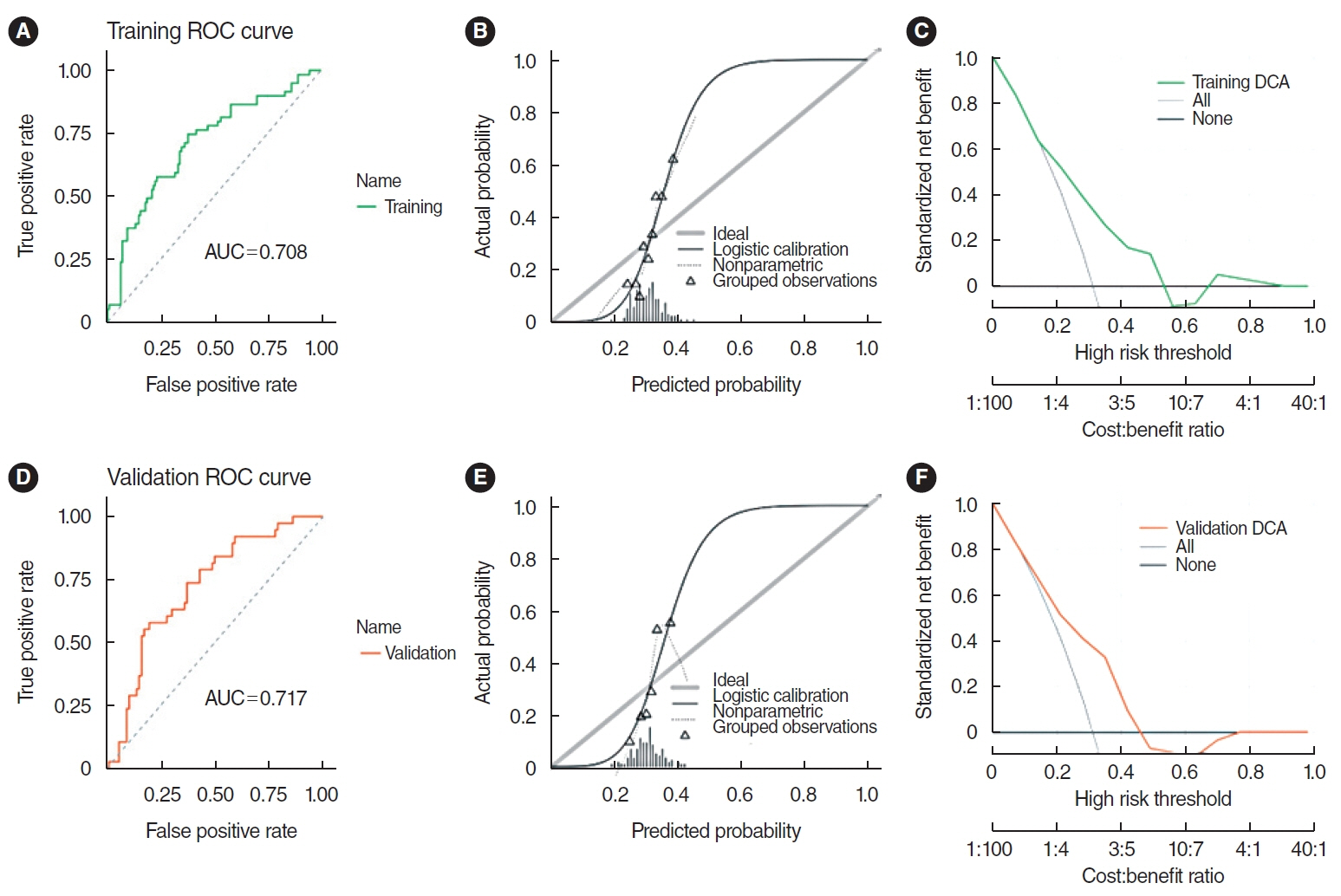
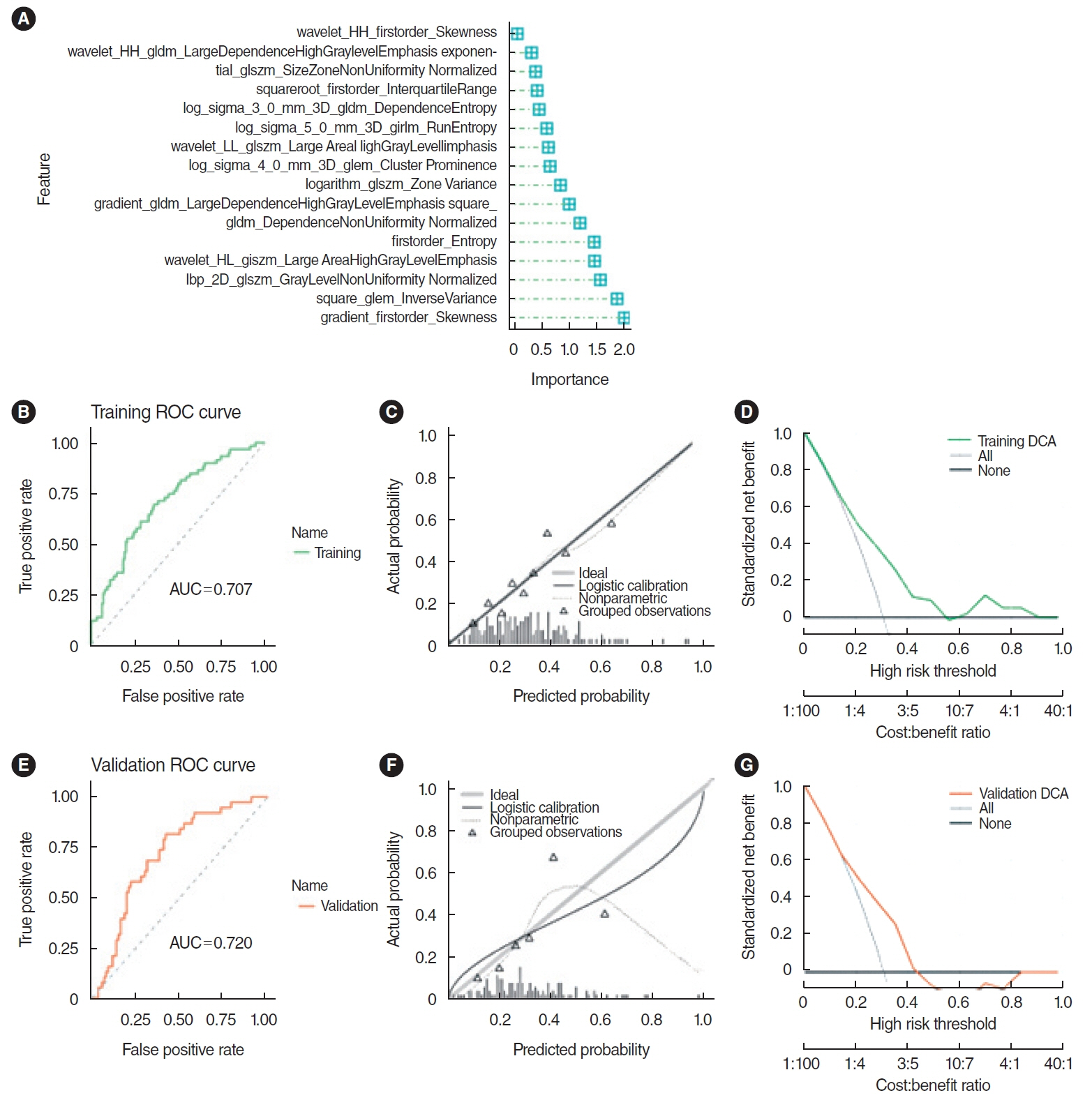
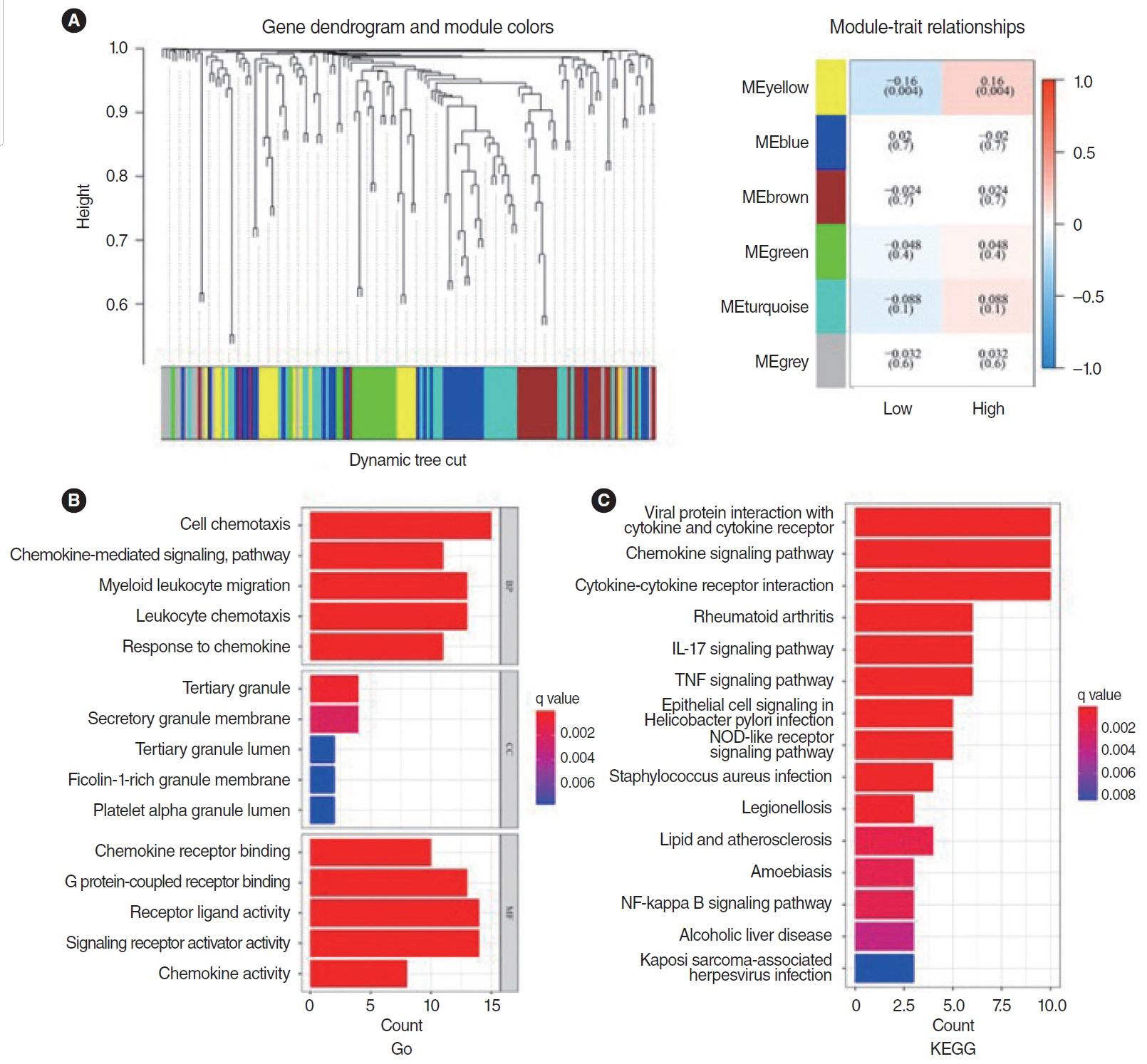
. Clinical information and transcripts per million mRNA sequencing data were obtained from The Cancer Genome Atlas (TCGA)-HNSCC dataset. We identified correlations between CXCL8 mRNA expression and patient survival rates using a Kaplan-Meier survival curve. A retrospective analysis of 313 samples diagnosed with HNSCC in the TCGA database was conducted. Pathomics features were extracted from hematoxylin and eosin–stained images, and then the minimum redundancy maximum relevance, with recursive feature elimination (mRMR-RFE) method was applied, followed by screening with the logistic regression algorithm.
Results
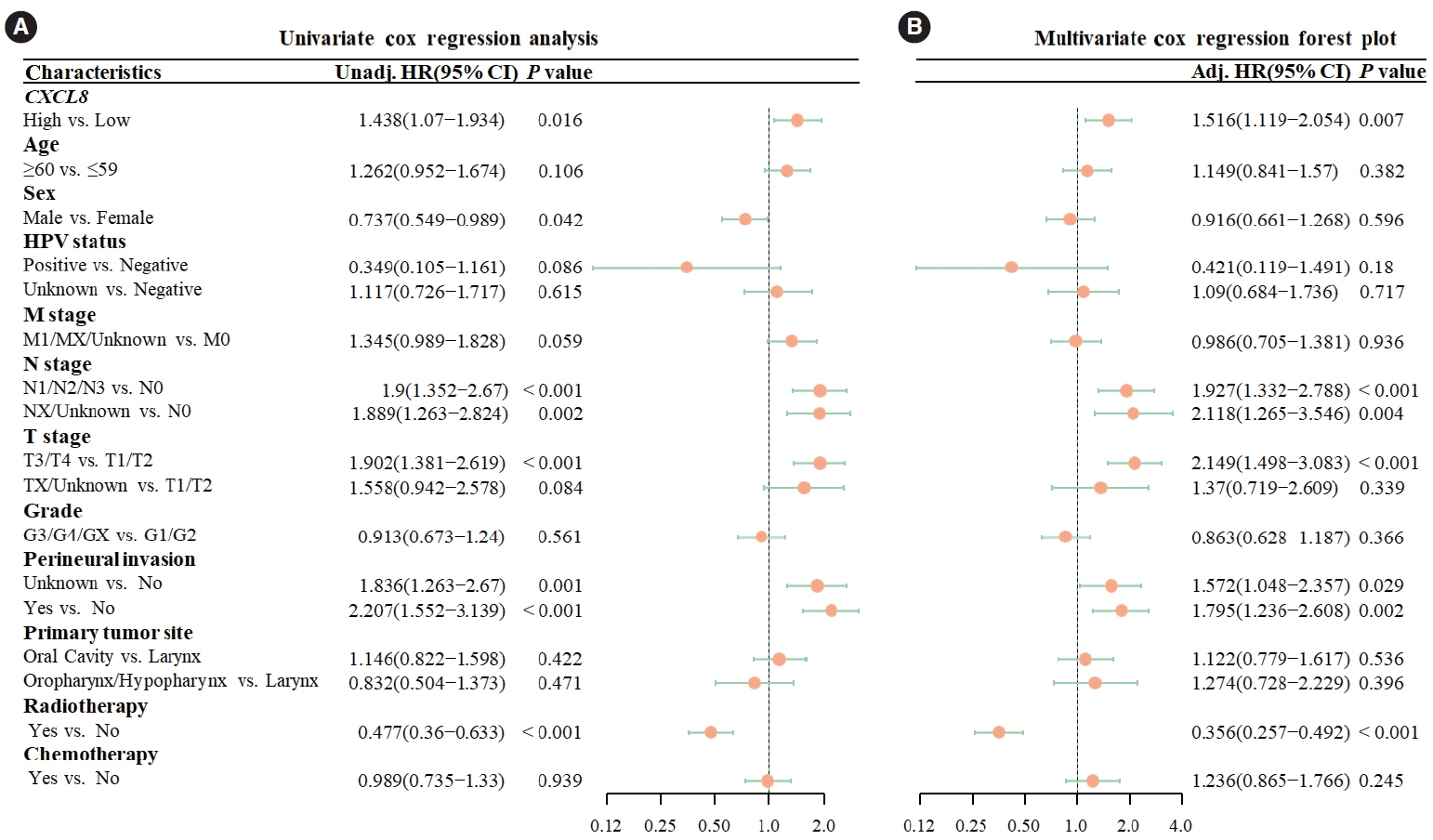
. Kaplan-Meier curves indicated that high expression of CXCL8 was significantly associated with decreased OS. The logistic regression pathomics model incorporated 16 radiomics features identified by the mRMR-RFE method in the training set and demonstrated strong performance in the testing set. Calibration plots showed that the probability of high gene expression predicted by the pathomics model was in good agreement with actual observations, suggesting the model’s high clinical applicability.
Conclusion
. The pathomics model of CXCL8 mRNA expression serves as an effective tool for predicting prognosis in patients with HNSCC and can aid in clinical decision-making. Elevated levels of CXCL8 expression may lead to reduced DNA damage and are associated with a pro-inflammatory tumor microenvironment, offering a potential therapeutic target.
Keyword
Figure
Reference
-
1. Xiao R, An Y, Ye W, Derakhshan A, Cheng H, Yang X, et al. Dual antagonist of cIAP/XIAP ASTX660 sensitizes HPV- and HPV+ head and neck cancers to TNFα, TRAIL, and radiation therapy. Clin Cancer Res. 2019; Nov. 25(21):6463–74.2. Weiss J, Sheth S, Deal AM, Grilley Olson JE, Patel S, Hackman TG, et al. Concurrent definitive immunoradiotherapy for patients with stage III-IV head and neck cancer and cisplatin contraindication. Clin Cancer Res. 2020; Aug. 26(16):4260–7.3. Karam SD, Reddy K, Blatchford PJ, Waxweiler T, DeLouize AM, Oweida A, et al. Final report of a phase I trial of Olaparib with cetuximab and radiation for heavy smoker patients with locally advanced head and neck cancer. Clin Cancer Res. 2018; Oct. 24(20):4949–59.4. Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers. Cancer. 2007; Oct. 110(7):1429–35.5. Ma Y, Wang B, He P, Qi W, Xiang L, Maswikiti EP, et al. Coagulation- and fibrinolysis-related genes for predicting survival and immunotherapy efficacy in colorectal cancer. Front Immunol. 2022; Nov. 13:1023908.6. Chen E, Qin X, Peng K, Xu X, Li W, Cheng X, et al. Identification of potential therapeutic targets among CXC chemokines in breast tumor microenvironment using integrative bioinformatics analysis. Cell Physiol Biochem. 2018; 45(5):1731–46.7. Qi WQ, Zhang Q, Wang JB. CXCL8 is a potential biomarker for predicting disease progression in gastric carcinoma. Transl Cancer Res. 2020; Feb. 9(2):1053–62.8. Wadapurkar RM, Sivaram A, Vyas R. RNA-Seq analysis of clinical samples from TCGA reveal molecular signatures for ovarian cancer. Cancer Invest. 2023; Apr. 41(4):394–404.9. Lukaszewicz-Zajac M, Paczek S, Muszynski P, Kozlowski M, Mroczko B. Comparison between clinical significance of serum CXCL-8 and classical tumor markers in oesophageal cancer (OC) patients. Clin Exp Med. 2019; May. 19(2):191–9.10. Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016; Oct. 31:61–71.11. Han ZJ, Li YB, Yang LX, Cheng HJ, Liu X, Chen H. Roles of the CXCL8-CXCR1/2 axis in the tumor microenvironment and immunotherapy. Molecules. 2021; Dec. 27(1):137.12. Classe M, Lerousseau M, Scoazec JY, Deutsch E. Perspectives in pathomics in head and neck cancer. Curr Opin Oncol. 2021; May. 33(3):175–83.13. Chen D, Lai J, Cheng J, Fu M, Lin L, Chen F, et al. Predicting peritoneal recurrence in gastric cancer with serosal invasion using a pathomics nomogram. iScience. 2023; Mar. 26(3):106246.14. Chen L, Zeng H, Zhang M, Luo Y, Ma X. Histopathological image and gene expression pattern analysis for predicting molecular features and prognosis of head and neck squamous cell carcinoma. Cancer Med. 2021; Jul. 10(13):4615–28.15. Zeng H, Chen L, Zhang M, Luo Y, Ma X. Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol Oncol. 2021; Oct. 163(1):171–80.16. Wang X, Chen H, Gan C, Lin H, Dou Q, Tsougenis E, et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans Cybern. 2020; Sep. 50(9):3950–62.17. Saednia K, Lagree A, Alera MA, Fleshner L, Shiner A, Law E, et al. Quantitative digital histopathology and machine learning to predict pathological complete response to chemotherapy in breast cancer patients using pre-treatment tumor biopsies. Sci Rep. 2022; Jun. 12(1):9690.18. Li H, Chen L, Zeng H, Liao Q, Ji J, Ma X. Integrative analysis of histopathological images and genomic data in colon adenocarcinoma. Front Oncol. 2021; Sep. 11:636451.19. Nishio M, Nishio M, Jimbo N, Nakane K. Homology-based image processing for automatic classification of histopathological images of lung tissue. Cancers (Basel). 2021; Mar. 13(6):1192.20. Huang Y, Wei L, Hu Y, Shao N, Lin Y, He S, et al. Multi-parametric MRI-based radiomics models for predicting molecular subtype and androgen receptor expression in breast cancer. Front Oncol. 2021; Aug. 11:706733.21. Xie J, Chen L, Tang Q, Wei W, Cao Y, Wu C, et al. A necroptosis-related prognostic model of uveal melanoma was constructed by single-cell sequencing analysis and weighted co-expression network analysis based on public databases. Front Immunol. 2022; Feb. 13:847624.22. Wang R, Dai W, Gong J, Huang M, Hu T, Li H, et al. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. 2022; Jan. 15(1):11.23. Chen D, Fu M, Chi L, Lin L, Cheng J, Xue W, et al. Prognostic and predictive value of a pathomics signature in gastric cancer. Nat Commun. 2022; Nov. 13(1):6903.24. Qu WF, Tian MX, Lu HW, Zhou YF, Liu WR, Tang Z, et al. Development of a deep pathomics score for predicting hepatocellular carcinoma recurrence after liver transplantation. Hepatol Int. 2023; Aug. 17(4):927–41.25. Sounbuli K, Mironova N, Alekseeva L. Diverse neutrophil functions in cancer and promising neutrophil-based cancer therapies. Int J Mol Sci. 2022; Dec. 23(24):15827.26. SenGupta S, Hein LE, Xu Y, Zhang J, Konwerski JR, Li Y, et al. Triple-negative breast cancer cells recruit neutrophils by secreting TGF-β and CXCR2 ligands. Front Immunol. 2021; Apr. 12:659996.27. Lin C, He H, Liu H, Li R, Chen Y, Qi Y, et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019; Oct. 68(10):1764–73.28. Yuen KC, Liu LF, Gupta V, Madireddi S, Keerthivasan S, Li C, et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat Med. 2020; May. 26(5):693–8.29. Li Y, Wu T, Gong S, Zhou H, Yu L, Liang M, et al. Analysis of the prognosis and therapeutic value of the CXC chemokine family in head and neck squamous cell carcinoma. Front Oncol. 2021; Jan. 10:570736.30. Chen X, Lei H, Cheng Y, Fang S, Sun W, Zhang X, et al. CXCL8, MMP12, and MMP13 are common biomarkers of periodontitis and oral squamous cell carcinoma. Oral Dis. 2022 Nov 2 [Epub]. https://doi.org/10.1111/odi.14419.31. Choi JH, Lee BS, Jang JY, Lee YS, Kim HJ, Roh J, et al. Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat Commun. 2023; Feb. 14(1):1055.32. Wang F, Zhang W, Chai Y, Wang H, Liu Z, He Y. Constrast-enhanced computed tomography radiomics predicts CD27 expression and clinical prognosis in head and neck squamous cell carcinoma. Front Immunol. 2022; Nov. 13:1015436.33. Fousek K, Horn LA, Palena C. Interleukin-8: a chemokine at the intersection of cancer plasticity, angiogenesis, and immune suppression. Pharmacol Ther. 2021; Mar. 219:107692.34. David JM, Dominguez C, Hamilton DH, Palena C. The IL-8/IL-8R axis: a double agent in tumor immune resistance. Vaccines (Basel). 2016; Jun. 4(3):22.35. Sanmamed MF, Perez-Gracia JL, Schalper KA, Fusco JP, Gonzalez A, Rodriguez-Ruiz ME, et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann Oncol. 2017; Aug. 28(8):1988–95.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Diagnostic Accuracy of Machine Learning Algorithms for Hepatitis A Antibody
- Development and Validation of a Prediction Model: Application to Digestive Cancer Research
- Machine Learning-Based Predictor for Treatment Outcomes of Patients With Salivary Gland Cancer After Operation
- Development and External Validation of a Machine Learning Model to Predict Pathological Complete Response After Neoadjuvant Chemotherapy in Breast Cancer
- Application of Machine Learning in Rhinology: A State of the Art Review