Lab Med Online.
2023 Oct;13(4):370-374. 10.47429/lmo.2023.13.4.370.
A Case of Chronic Myeloid Leukemia with Micro BCR::ABL1 Rearrangement: Precaution in Reverse Transcription PCR to Prevent False Negativity
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University College of Medicine, Severance Hospital, Seoul
- 2Division of Hematology, Department of Internal Medicine, Yonsei University College of Medicine, Severance Hospital, Seoul, Korea
- KMID: 2552765
- DOI: http://doi.org/10.47429/lmo.2023.13.4.370
Abstract
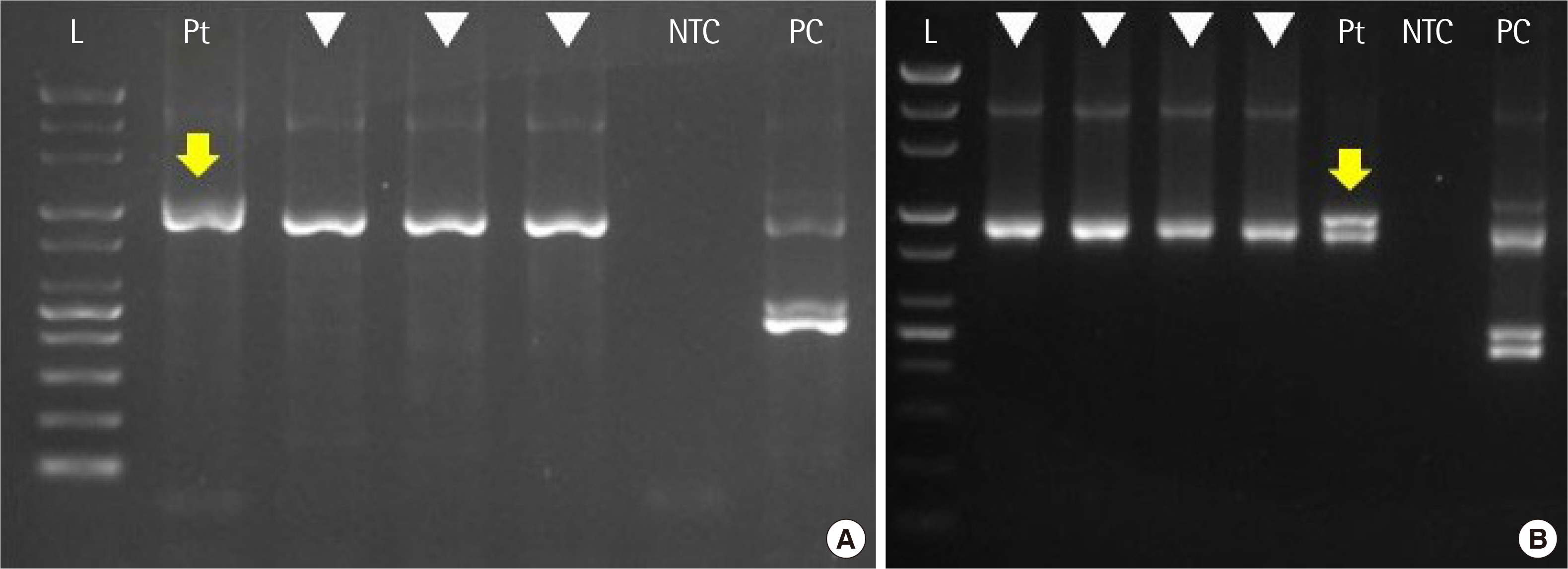
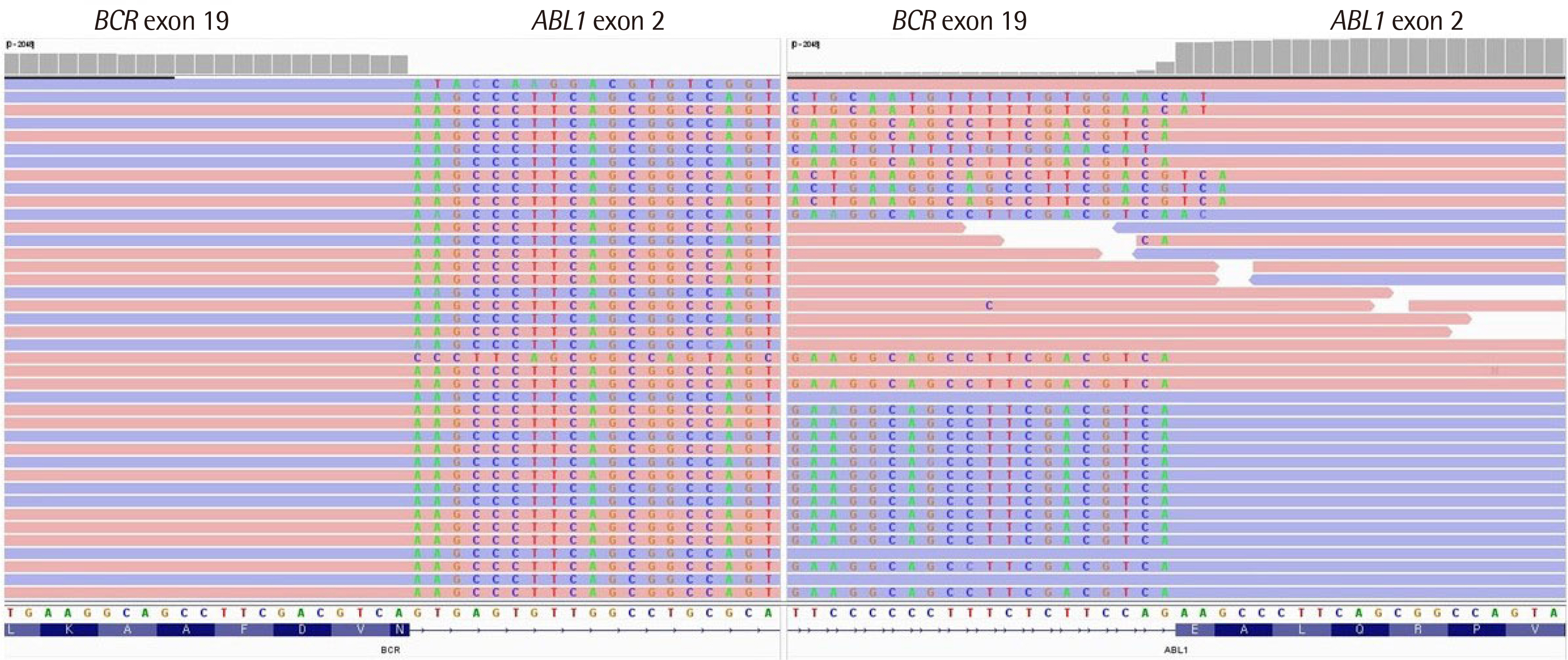
- We report a patient negative for BCR::ABL1 in qualitative reverse transcription (RT)-PCR but subsequently reported to be positive for t(9;22) (q34;q11.2) in conventional karyotyping. The patient was finally diagnosed with micro-type chronic myeloid leukemia after re-examining RT-PCR and performing targeted RNA sequencing. Through this case, we highlight the risk of false negativity when interpreting RT-PCR to detect microtype fusion. Upon re-examining RT-PCR results, the patient’s internal control band was thicker than others. After extending the electrophoresis run time, a 911-bp internal control band and a target band around the level of 1.0 kb were separated. We confirmed a fusion breakpoint (BCR exon 19 and ABL1 exon 2) by targeted RNA sequencing, and it corresponds to 1,012 bp-sized e19a2 (c3a2) type among four micro-type fusion transcripts that RT-PCR HemaVision ® kit M6B can detect.
Figure
Reference
-
1. Melo JV. 1996; The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 88:2375–84. DOI: 10.1182/blood.V88.7.2375.bloodjournal8872375. PMID: 8839828.2. Ben-Neriah Y, Daley GQ, Mes-Masson AM, Witte ON, Baltimore D. 1986; The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science. 233:212–4. DOI: 10.1126/science.3460176. PMID: 3460176.3. Ohsaka A, Shiina S, Kobayashi M, Kudo H, Kawaguchi R. 2002; Philadelphia chromosome-positive chronic myeloid leukemia expressing p190(BCR-ABL). Intern Med. 41:1183–7. DOI: 10.2169/internalmedicine.41.1183. PMID: 12521212.4. Komorowski L, Fidyt K, Patkowska E, Firczuk M. 2020; Philadelphia chromosome-positive leukemia in the lymphoid lineage-similarities and differences with the myeloid lineage and specific vulnerabilities. Int J Mol Sci. 21:5776. DOI: 10.3390/ijms21165776. PMID: 32806528. PMCID: PMC7460962.5. Ito T, Tanaka H, Tanaka K, Ito K, Kyo T, Dohy H, et al. 2004; Insertion of a genomic fragment of chromosome 19 between BCR intron 19 and ABL intron 1a in a chronic myeloid leukaemia patient with micro-BCR-ABL (e19a2) transcript. Br J Haematol. 126:752–3. DOI: 10.1111/j.1365-2141.2004.05119.x. PMID: 15327531.6. Mondal BC, Majumdar S, Dasgupta UB, Chaudhuri U, Chakrabarti P, Bhattacharyya S. 2006; e19a2 BCR-ABL fusion transcript in typical chronic myeloid leukaemia: a report of two cases. J Clin Pathol. 59:1102–3. DOI: 10.1136/jcp.2005.029595. PMID: 17021137. PMCID: PMC1861751.7. Haskovec C, Ponzetto C, Polák J, Maritano D, Zemanová Z, Serra A, et al. 1998; P230 BCR/ABL protein may be associated with an acute leukaemia phenotype. Br J Haematol. 103:1104–8. DOI: 10.1046/j.1365-2141.1998.01098.x. PMID: 9886327.8. Kim HY, Park S, Kim SH, Kim HJ. 2017; The first report of chronic myelogenous leukemia with e19a2 micro-BCR/ABL1 presenting with massive myelofibrosis. Blood Cells Mol Dis. 65:68–70. DOI: 10.1016/j.bcmd.2017.05.001. PMID: 28571780.9. Lee J, Kim DS, Lee HS, Choi SI, Cho YG. 2017; Concurrence of e1a2 and e19a2 BCR-ABL1 Ffusion Ttranscripts in a Tyypical Ccase of Cchronic Mmyeloid Lleukemia. Ann Lab Med. 37:74–6. DOI: 10.3343/alm.2017.37.1.74. PMID: 27834071. PMCID: PMC5107623.10. Kim JH, Lee WM, Ryoo NH, Ha JS, Jeon DS, Kim JR, et al. 2008; A case of atypical CML with micro BCR/ABL rearrangement. Korean J Hematol. 43:184–9. DOI: 10.5045/kjh.2008.43.3.184.11. Goh HG, Hwang JY, Kim SH, Lee YH, Kim YL, Kim DW. 2006; Comprehensive analysis of BCR-ABL transcript types in Korean CML patients using a newly developed multiplex RT-PCR. Transl Res. 148:249–56. DOI: 10.1016/j.trsl.2006.07.002. PMID: 17145570.12. Molica M, Abruzzese E, Breccia M. 2020; Prognostic significance of transcript-type BCR-ABL1 in chronic myeloid leukemia. Mediterr J Hematol Infect Dis. 12:e2020062. DOI: 10.4084/mjhid.2020.062. PMID: 32952973. PMCID: PMC7485470.13. Engvall M, Cahill N, Jonsson BI, Höglund M, Hallböök H, Cavelier L. 2020; Detection of leukemia gene fusions by targeted RNA-sequencing in routine diagnostics. BMC Med Genomics. 13:106. DOI: 10.1186/s12920-020-00739-4. PMID: 32727569. PMCID: PMC7388219.14. Langabeer SE. 2015; Standardized molecular monitoring for variant BCR-ABL1 transcripts in chronic myeloid leukemia. Arch Pathol Lab Med. 139:969. DOI: 10.5858/arpa.2014-0522-LE. PMID: 26230589.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of Acute Promyelocytic Leukemia with Co-existence of BCR-ABL1 and PML-RARA Rearrangements Detected by PCR
- A Case of Concomitant Inv(3)(q21q26) and Cryptic BCR/ABL1 Rearrangement in the Blast Crisis of Chronic Myeloid Leukemia
- A Case of Atypical Chronic Myeloid Leukemia with the JAK2V617F Mutation
- detection of BCR-ABL gene rearrangement by use of the polymerase chain reaction in chronic myeloid leukemia
- Analysis of BCR-ABL Fusion Transcripts of Chronic Myeloid Leukemia Patients in Korea