Korean J Gastroenterol.
2024 Jan;83(1):6-16. 10.4166/kjg.2023.132.
Transcription Silencing and CpGs Hypermethylation as Therapeutic Gene Editing in Clinical Colorectal Adenocarcinoma Repression
- Affiliations
-
- 1Department of Internal Medicine, Ninevah University, College of Medicine, Mosul, Iraq
- KMID: 2552237
- DOI: http://doi.org/10.4166/kjg.2023.132
Abstract
- Background/Aims
Colorectal cancer is the most common cancer in oncopathology, with an increasing incidence among the elderly during the last decade. Various genetic and environmental factors play important roles in the emergence of colorectal adenocarcinoma. Non-coding RNAs, approximately 20–22 nucleotides, are transcribed irregularly in many cancer cells and play a critical role in many metabolic pathways in clinical cancer cases. DNA methylation is a critical epigenetic alteration that controls gene expression. In the current study, transcriptional silencing and CpG hypermethylation were developed as a therapeutic gene editing strategy for the clinical repression of colorectal adenocarcinoma.
Methods
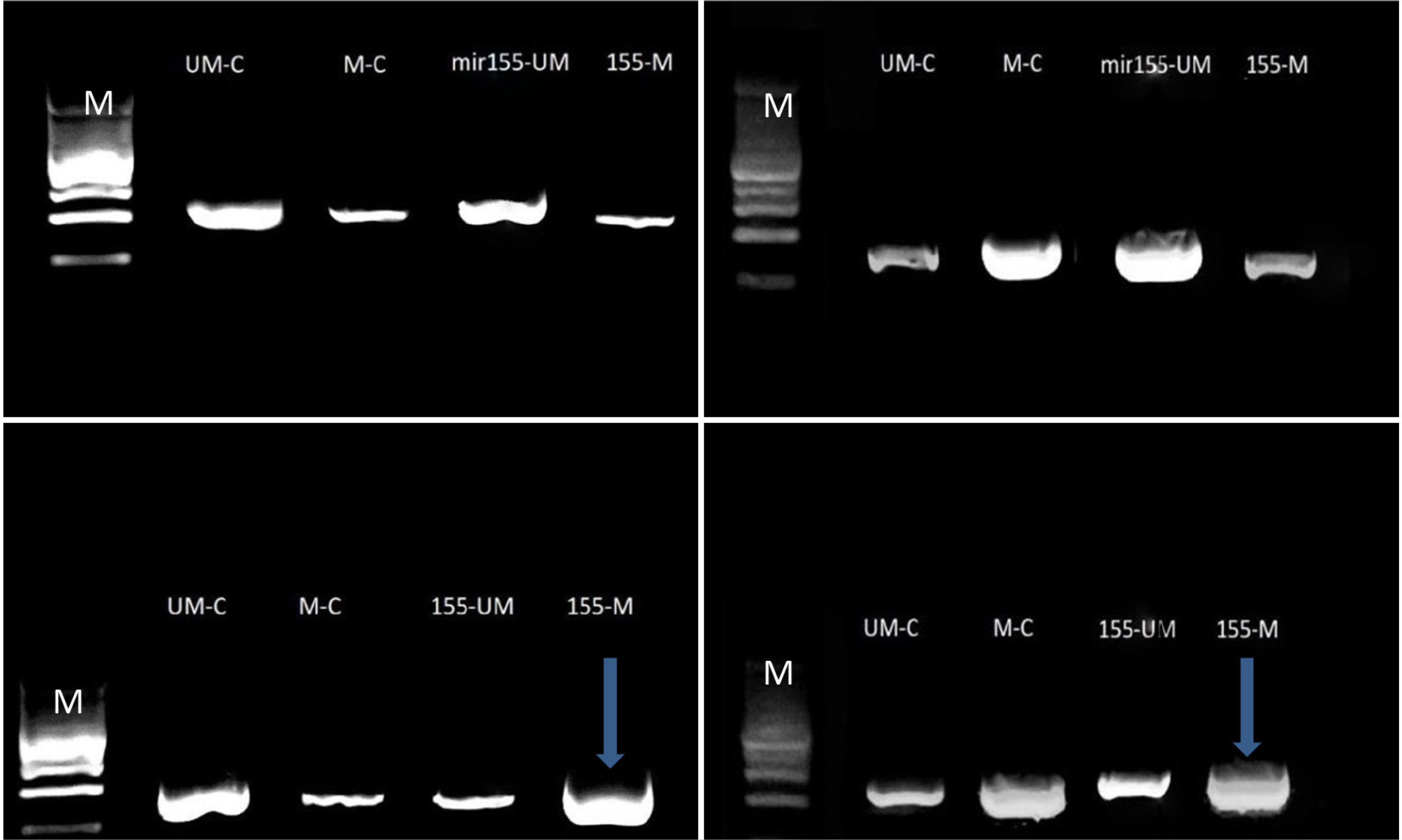
A human colorectal adenocarcinoma cell line (Caco2) and a normal lung fibroblast cell line (Wi38) were utilized as the paradigms in this research to examine the effect of mir155 molecule transfection and CpGs-island (CGI) methylation. Cell counting was achieved using six-well and 24-well plates before transfection using a hemocytometer. The two cell lines were transfected with the mir155 agomir and antagomir molecules. The transfection efficiency, cell viability, cell IC 50 , and target gene expression were measured, and CGIs-methylation was achieved by bisulfate conversion.
Results
The outcomes revealed the downregulation of oncogenes (AKT1 and VCAM1 genes as cancer-associated genes) and the upregulation of tumor suppressor genes (TSGs, Tp53 and KEAP1). In addition, CpG-islands methylation showed significant blocking of the oncogene promoter regions, and the switch on of TSG promoter regions was continuous.
Conclusions
miRNA-CGI-methylation led to the regression of Caco2 cell proliferation, suggesting the potential use of RNA silencing and DNA methylation in targeted gene therapy for colorectal cancer. (
Keyword
Figure
Reference
-
1. Schreuders EH, Ruco A, Rabeneck L, et al. 2015; Colorectal cancer screening: a global overview of existing programmes. Gut. 64:1637–1649. DOI: 10.1136/gutjnl-2014-309086. PMID: 26041752.2. Duggan MA, Anderson WF, Altekruse S, Penberthy L, Sherman ME. 2016; The surveillance, epidemiology, and end results (SEER) program and pathology: Toward strengthening the critical relationship. Am J Surg Pathol. 40:e94–e102. DOI: 10.1097/PAS.0000000000000749. PMID: 27740970. PMCID: PMC5106320.3. Jonas S, Izaurralde E. 2015; Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 16:421–433. DOI: 10.1038/nrg3965. PMID: 26077373.4. Kim YK, Kim B, Kim VN. 2016; Re-evaluation of the roles of DROSHA, Export in 5, and DICER in microRNA biogenesis. Proc Natl Acad Sci U S A. 113:E1881–E1889. DOI: 10.1073/pnas.1602532113. PMID: 26976605. PMCID: PMC4822641.5. Chan JJ, Tay Y. 2018; Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 19:1310. DOI: 10.3390/ijms19051310. PMID: 29702599. PMCID: PMC5983611.6. Alexandrov LB, Nik-Zainal S, Wedge DC, et al. 2013; Signatures of mutational processes in human cancer. Nature. 500:415–421. DOI: 10.1038/nature12477. PMID: 23945592. PMCID: PMC3776390.7. Gao X, Wang N, Wu S, Cui H, An X, Yang Y. 2019; Long non‑coding RNA FER1L4 inhibits cell proliferation and metastasis through regulation of the PI3K/AKT signaling pathway in lung cancer cells. Mol Med Rep. 20:182–190. DOI: 10.3892/mmr.2019.10219. PMID: 31115514. PMCID: PMC6579969.8. Rice GE, Bevilacqua MP. 1989; An inducible endothelial cell surface glycoprotein mediates melanoma adhesion. Science. 246:1303–1306. DOI: 10.1126/science.2588007. PMID: 2588007.9. Dolgin E. 2017; The most popular genes in the human genome. Nature. 551:427–431. DOI: 10.1038/d41586-017-07291-9. PMID: 29168817.10. Chen Q, Zhang XH, Massagué J. 2011; Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 20:538–549. DOI: 10.1016/j.ccr.2011.08.025. PMID: 22014578. PMCID: PMC3293160.11. Mello SS, Attardi LD. 2018; Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 51:65–72. DOI: 10.1016/j.ceb.2017.11.005. PMID: 29195118. PMCID: PMC5949255.12. Parrales A, Iwakuma T. 2015; Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 5:288. DOI: 10.3389/fonc.2015.00288. PMID: 26732534. PMCID: PMC4685147.13. Sadeghi MR, Jeddi F, Soozangar N, Somi MH, Samadi N. 2017; The role of Nrf2-Keap1 axis in colorectal cancer, progression, and chemoresistance. Tumour Biol. 39:1010428317705510. DOI: 10.1177/1010428317705510. PMID: 28621229.14. Schübeler D. 2015; ESCI award lecture: regulation, function and biomarker potential of DNA methylation. Eur J Clin Invest. 45:288–293. DOI: 10.1111/eci.12403. PMID: 25608229.15. Mosca L, Vitiello F, Borzacchiello L, et al. 2021; Mutual correlation between non-coding RNA and S-Adenosylmethionine in human cancer: Roles and therapeutic opportunities. Cancers (Basel). 13:3264. DOI: 10.3390/cancers13133264. PMID: 34209866. PMCID: PMC8268931.16. Pajares MJ, Alemany-Cosme E, Goñi S, Bandres E, Palanca-Ballester C, Sandoval J. 2021; Epigenetic regulation of microRNAs in cancer: Shortening the distance from bench to bedside. Int J Mol Sci. 22:7350. DOI: 10.3390/ijms22147350. PMID: 34298969. PMCID: PMC8306710.17. Livak KJ, Schmittgen TD. 2001; Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 25:402–408. DOI: 10.1006/meth.2001.1262. PMID: 11846609.18. Li N, Cui T, Guo W, Wang D, Mao L. 2019; MiR-155-5p accelerates the metastasis of cervical cancer cell via targeting TP53INP1. Onco Targets Ther. 12:3181–3196. DOI: 10.2147/OTT.S193097. PMID: 31118671. PMCID: PMC6500876.19. Dong Y, Yu T, Ding L, et al. 2018; A dual targeting dendrimer-mediated siRNA delivery system for effective gene silencing in cancer therapy. J Am Chem Soc. 140:16264–16274. DOI: 10.1021/jacs.8b10021. PMID: 30346764.20. He B, Gao SQ, Huang LD, et al. 2015; MicroRNA-155 promotes the proliferation and invasion abilities of colon cancer cells by targeting quaking. Mol Med Rep. 11:2355–2359. DOI: 10.3892/mmr.2014.2994. PMID: 25420938.21. Shit MN, Al-Hamadani AH, Al-Askeri MA. Epigenetic regulation of colorectal adenocarcinoma cells using ncRNA [PhD thesis]. University of Al-Qadisiyah College of Medicine;Al-Qādisiyyah:22. Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. 2013; Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 31:1375–1380. DOI: 10.3892/ijmm.2013.1348. PMID: 23588589.23. Liu N, Jiang F, Han XY, et al. 2018; MiRNA-155 promotes the invasion of colorectal cancer SW-480 cells through regulating the Wnt/β-catenin. Eur Rev Med Pharmacol Sci. 22:101–109.24. Colemon A, Harris TM, Ramanathan S. 2020; DNA hypomethylation drives changes in MAGE-A gene expression resulting in alteration of proliferative status of cells. Genes Environ. 42:24. DOI: 10.1186/s41021-020-00162-2. PMID: 32760472. PMCID: PMC7392716.25. Lee YT, Tan YJ, Oon CE. 2018; Molecular targeted therapy: Treating cancer with specificity. Eur J Pharmacol. 834:188–196. DOI: 10.1016/j.ejphar.2018.07.034. PMID: 30031797.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Epigenetic Changes (Aberrant DNA Methylation) in Colorectal Neoplasia
- The upstream sequence of Mycobacterium leprae 18-kDa gene confers transcription repression activity in orientation-independent manner
- Heterologous Regulation of BCG hsp65 Promoter by M. leprae 18 kDa Transcription Repression Responsive Element
- Quantitative Analysis of Cancer-associated Gene Methylation Connected to Risk Factors in Korean Colorectal Cancer Patients
- Epigenetic Alterations and Loss of Imprinting in Colorectal Cancer