Ann Lab Med.
2023 Nov;43(6):638-641. 10.3343/alm.2023.43.6.638.
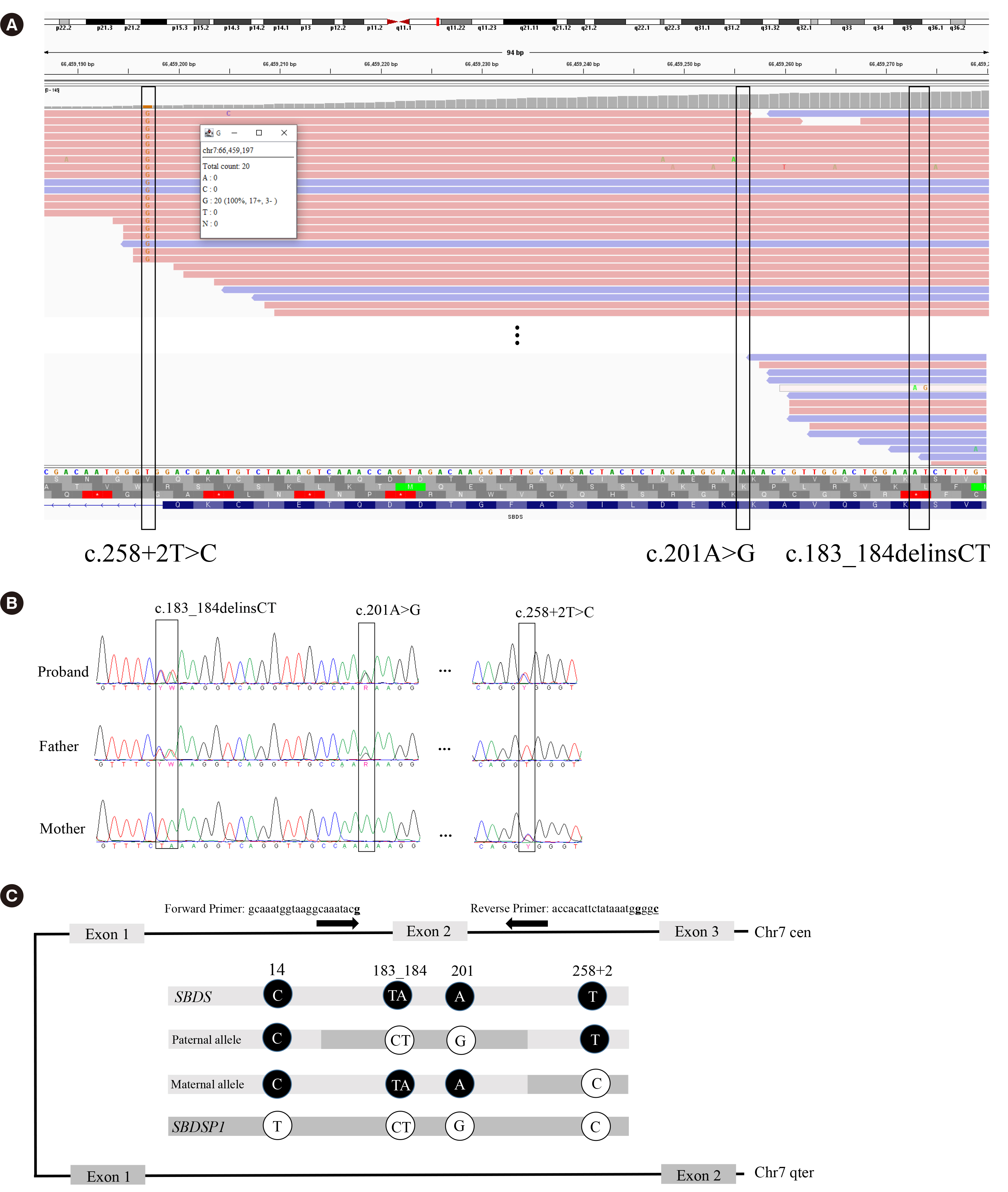
Variant Allele Frequency of Pseudogene-Related Variants in Short-read Next-Generation Sequencing Data May Mislead Genetic Diagnosis: A Case of Shwachman-Diamond Syndrome
- Affiliations
-
- 1Department of Laboratory Medicine, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Pediatrics, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 3Rare Disease Center, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- 4Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2552045
- DOI: http://doi.org/10.3343/alm.2023.43.6.638
Figure
Reference
-
1. Nelson A, Myers K. Adam MP, Mirzaa GM, editors. 2008. Shwachman-Diamond Syndrome. GeneReviews®. University of Washington, Seattle;Seattle:2. Furutani E, Liu S, Galvin A, Steltz S, Malsch MM, Loveless SK, et al. 2022; Hematologic complications with age in Shwachman-Diamond syndrome. Blood Adv. 6:297–306. DOI: 10.1182/bloodadvances.2021005539. PMID: 34758064. PMCID: PMC8753194.
Article3. Orphanet. The portal for rare diseases and orphan drugs. https://www.orpha.net/. Updated on June 2014.4. Warren AJ. 2018; Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Adv Biol Regul. 67:109–27. DOI: 10.1016/j.jbior.2017.09.002. PMID: 28942353. PMCID: PMC6710477.
Article5. Lawal OS, Mathur N, Eapi S, Chowdhury R, Malik BH. 2020; Liver and cardiac involvement in Shwachman-Diamond syndrome: a literature review. Cureus. 12:e6676. DOI: 10.7759/cureus.6676.
Article6. Yamada M, Uehara T, Suzuki H, Takenouchi T, Inui A, Ikemiyagi M, et al. 2020; Shortfall of exome analysis for diagnosis of Shwachman-Diamond syndrome: mismapping due to the pseudogene SBDSP1. Am J Med Genet A. 182:1631–6. DOI: 10.1002/ajmg.a.61598. PMID: 32412173.7. Wu D, Zhang L, Qiang Y, Wang K. 2022; Improved detection of SBDS gene mutation by a new method of next-generation sequencing analysis based on the Chinese mutation spectrum. PLoS One. 17:e0269029. DOI: 10.1371/journal.pone.0269029. PMID: 36512530. PMCID: PMC9747038.
Article8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article9. Peng X, Dong X, Wang Y, Wu B, Wang H, Lu W, et al. 2022; Overcoming the pitfalls of next-generation sequencing-based molecular diagnosis of Shwachman-Diamond syndrome. J Mol Diagn. 24:1240–53. DOI: 10.1016/j.jmoldx.2022.09.002. PMID: 36162759.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetics and genomics of bone marrow failure syndrome
- A Case of Shwachman-Diamond Syndrome
- A Case of Shwachman-Diamond Syndrome Confirmed with Genetic Analysis in a Korean Child
- Long-read sequencing of 12 samples discovered novel variants within human leukocyte antigen region
- Clinical applications of next-generation sequencing in the diagnosis of genetic disorders in Korea: a narrative review