Ann Lab Med.
2023 Jul;43(4):355-363. 10.3343/alm.2023.43.4.355.
Development of a Single-nucleotide Polymorphism Genotyping Assay for the Rapid Detection of Vancomycin-intermediate Resistance in Staphylococcus aureus Epidemic Lineage ST5
- Affiliations
-
- 1Division of Antimicrobial Resistance Research, Center for Infectious Diseases Research, National Institute of Health, Korea Disease Control and Prevention Agency, Cheongju, Korea
- 2Division of Zoonotic and Vector Borne Disease Research, Center for Infectious Diseases Research, National Institute of Health, Korea Disease Control and Prevention Agency, Cheongju, Korea
- KMID: 2551953
- DOI: http://doi.org/10.3343/alm.2023.43.4.355
Abstract
- Background
Vancomycin is a treatment option for patients with severe methicillin-resistant Staphylococcus aureus (MRSA) infection. Unfortunately, reduced susceptibility to vancomycin in S. aureus is becoming increasingly common. We developed a method for the rapid and accurate diagnosis of vancomycin-intermediate resistant S. aureus (VISA).
Methods
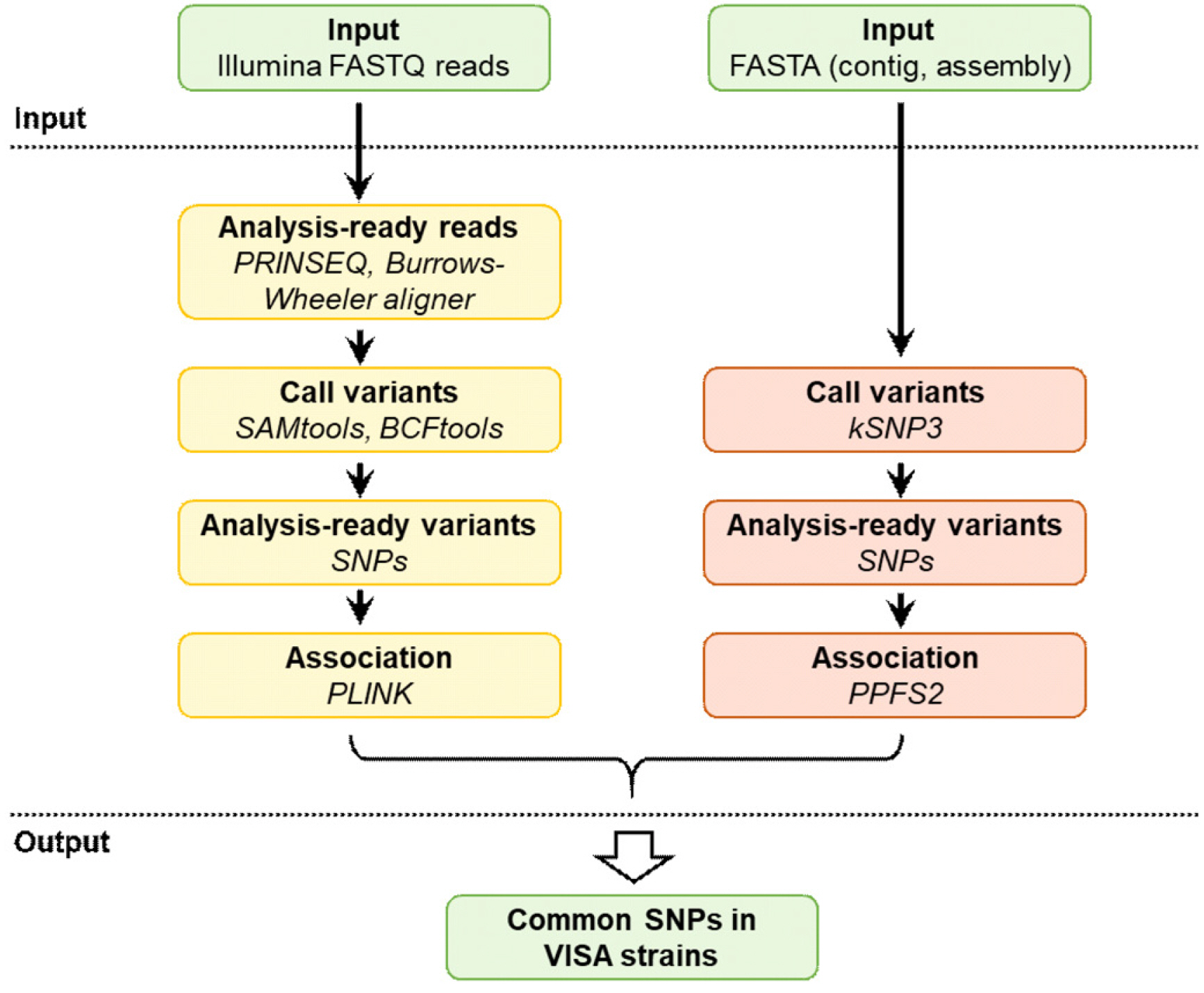
We performed a microbial genome-wide association study to discriminate between VISA and vancomycin-susceptible S. aureus (VSSA) using 42 whole-genome sequences. A TaqMan single-nucleotide polymorphism (SNP) genotyping assay was developed to detect target SNPs in VISA strains.
Results
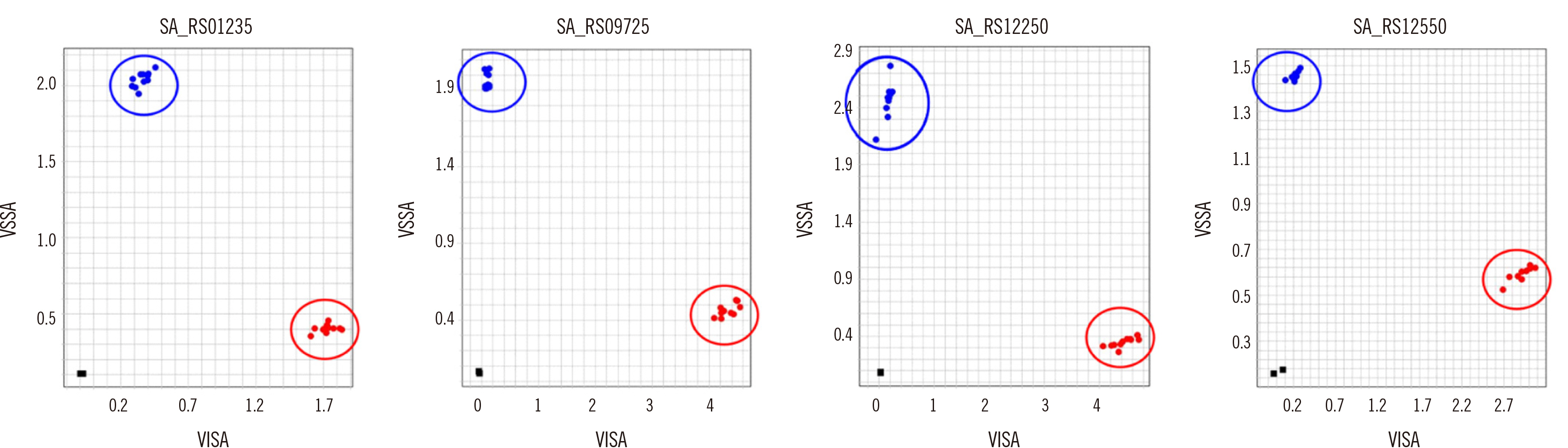
Four SNPs in the VISA strains resulting in nonsynonymous amino-acid substitutions were associated with reduced susceptibility to vancomycin: SA_RS01235 (G203S), SA_RS09725 (V171A), SA_RS12250 (I48F), and SA_RS12550 (G478A). These four SNPs were mainly detected in the typical hospital-associated sequence type (ST)5 clonal lineage. The TaqMan assay successfully detected all four SNPs using as little as 0.2 ng DNA per reaction. Using 10 VSSA and VISA clinical strains each, we validated that the assay accurately discriminates between VISA and VSSA.
Conclusions
The TaqMan SNP genotyping assay targeting four SNPs may be an alternative to current standard methods for the rapid detection of vancomycin-intermediate resistance in S. aureus epidemic lineage ST5.
Keyword
Figure
Reference
-
1. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. 2019; Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 17:203–18. DOI: 10.1038/s41579-018-0147-4. PMID: 30737488. PMCID: PMC6939889.
Article2. Chambers HF, DeLeo FR. 2009; Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 7:629–41. DOI: 10.1038/nrmicro2200. PMID: 19680247. PMCID: PMC2871281.
Article3. Tenover FC, Biddle JW, Lancaster MV. 2001; Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg Infect Dis. 7:327–32. DOI: 10.3201/eid0702.010237. PMID: 11294734. PMCID: PMC2631729.4. Tenover FC, Moellering RC. 2007; The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis. 44:1208–15. DOI: 10.1086/513203. PMID: 17407040.5. Cong Y, Yang S, Rao X. 2019; Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res. 21:169–76. DOI: 10.1016/j.jare.2019.10.005. PMID: 32071785. PMCID: PMC7015472.6. de Niederhäusern S, Bondi M, Messi P, Iseppi R, Sabia C, Manicardi G, et al. 2011; Vancomycin-resistance transferability from VanA enterococci to Staphylococcus aureus. Curr Microbiol. 62:1363–7. DOI: 10.1007/s00284-011-9868-6. PMID: 21234755.
Article7. Hafer C, Lin Y, Kornblum J, Lowy FD, Uhlemann AC. 2012; Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 56:5845–51. DOI: 10.1128/AAC.01139-12. PMID: 22948864. PMCID: PMC3486570.
Article8. Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, et al. 2007; Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 104:9451–6. DOI: 10.1073/pnas.0609839104. PMID: 17517606. PMCID: PMC1890515.9. Vidaillac C, Gardete S, Tewhey R, Sakoulas G, Kaatz GW, Rose WE, et al. 2013; Alternative mutational pathways to intermediate resistance to vancomycin in methicillin-resistant Staphylococcus aureus. J Infect Dis. 208:67–74. DOI: 10.1093/infdis/jit127. PMID: 23539745. PMCID: PMC3666135.
Article10. Howden BP, Peleg AY, Stinear TP. 2014; The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect Genet Evol. 21:575–82. DOI: 10.1016/j.meegid.2013.03.047. PMID: 23567819.11. Hu Q, Peng H, Rao X. 2016; Molecular events for promotion of vancomycin resistance in vancomycin intermediate Staphylococcus aureus. Front Microbiol. 7:1601. DOI: 10.3389/fmicb.2016.01601. PMID: 27790199. PMCID: PMC5062060.12. Phillips CJ, Wells NA, Martinello M, Smith S, Woodman RJ, Gordon DL. 2016; Optimizing the detection of methicillin-resistant Staphylococcus aureus with elevated vancomycin minimum inhibitory concentrations within the susceptible range. Infect Drug Resist. 9:87–92. DOI: 10.2147/IDR.S107961. PMID: 27330319. PMCID: PMC4898034.13. Howden BP, Davies JK, Johnson PD, Stinear TP, Grayson ML. 2010; Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin Microbiol Rev. 23:99–139. DOI: 10.1128/CMR.00042-09. PMID: 20065327. PMCID: PMC2806658.
Article14. CLSI. 2016. Performance standards for antimicrobial susceptibility testing. 26th ed. CLSI M100S. Clinical and Laboratory Standards Institute;Wayne, PA:15. Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007; Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 23:673–9. DOI: 10.1093/bioinformatics/btm009. PMID: 17237039. PMCID: PMC2387122.
Article16. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. 2014; The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42(Database issue):D206–14. DOI: 10.1093/nar/gkt1226. PMID: 24293654. PMCID: PMC3965101.
Article17. Schmieder R, Edwards R. 2011; Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27:863–4. DOI: 10.1093/bioinformatics/btr026. PMID: 21278185. PMCID: PMC3051327.
Article18. Li H, Durbin R. 2010; Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–95. DOI: 10.1093/bioinformatics/btp698. PMID: 20080505. PMCID: PMC2828108.
Article19. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. 2007; PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–75. DOI: 10.1086/519795. PMID: 17701901. PMCID: PMC1950838.
Article20. Hall BG. 2014; SNP-associations and phenotype predictions from hundreds of microbial genomes without genome alignments. PLoS One. 9:e90490. DOI: 10.1371/journal.pone.0090490. PMID: 24587377. PMCID: PMC3938750.
Article21. Gardner SN, Slezak T, Hall BG. 2015; kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 31:2877–8. DOI: 10.1093/bioinformatics/btv271. PMID: 25913206.
Article22. Broccanello C, Chiodi C, Funk A, McGrath JM, Panella L, Stevanato P. 2018; Comparison of three PCR-based assays for SNP genotyping in plants. Plant Methods. 14:28. DOI: 10.1186/s13007-018-0295-6. PMID: 29610576. PMCID: PMC5872507.
Article23. Lees JA, Croucher NJ, Goldblatt D, Nosten F, Parkhill J, Turner C, et al. 2017; Genome-wide identification of lineage and locus specific variation associated with pneumococcal carriage duration. Elife. 6:e26255. DOI: 10.7554/eLife.26255. PMID: 28742023. PMCID: PMC5576492.
Article24. Laabei M, Recker M, Rudkin JK, Aldeljawi M, Gulay Z, Sloan TJ, et al. 2014; Predicting the virulence of MRSA from its genome sequence. Genome Res. 24:839–49. DOI: 10.1101/gr.165415.113. PMID: 24717264. PMCID: PMC4009613.
Article25. Alam MT, Petit RA 3rd, Crispell EK, Thornton TA, Conneely KN, Jiang Y, et al. 2014; Dissecting vancomycin-intermediate resistance in Staphylococcus aureus using genome-wide association. Genome Biol Evol. 6:1174–85. DOI: 10.1093/gbe/evu092. PMID: 24787619. PMCID: PMC4040999.26. Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. 2013; Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 45:1183–9. DOI: 10.1038/ng.2747. PMID: 23995135. PMCID: PMC3887553.
Article27. Earle SG, Wu CH, Charlesworth J, Stoesser N, Gordon NC, Walker TM, et al. 2016; Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol. 1:16041. DOI: 10.1038/nmicrobiol.2016.41. PMID: 27572646. PMCID: PMC5049680.
Article28. Gardner SG, Marshall DD, Daum RS, Powers R, Somerville GA. 2017; Metabolic mitigation of Staphylococcus aureus vancomycin intermediate-level susceptibility. Antimicrob Agents Chemother. 62:e01608–17. DOI: 10.1128/AAC.01608-17. PMID: 29109158. PMCID: PMC5740343.29. Alexander EL, Gardete S, Bar HY, Wells MT, Tomasz A, Rhee KY. 2014; Intermediate-type vancomycin resistance (VISA) in genetically-distinct Staphylococcus aureus isolates is linked to specific, reversible metabolic alterations. PLoS One. 9:e97137. DOI: 10.1371/journal.pone.0097137. PMID: 24817125. PMCID: PMC4016254.30. Nelson JL, Rice KC, Slater SR, Fox PM, Archer GL, Bayles KW, et al. 2007; Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob Agents Chemother. 51:616–22. DOI: 10.1128/AAC.01057-06. PMID: 17130298. PMCID: PMC1797750.
Article31. Zhang S, Sun X, Chang W, Dai Y, Ma X. 2015; Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS One. 10:e0136082. DOI: 10.1371/journal.pone.0136082. PMID: 26287490. PMCID: PMC4546009.32. Park C, Rho K, Shin J, Cho SY, Lee DG, Chung YJ. 2021; Genomic analysis of heterogeneous vancomycin-intermediate Staphylococcus aureus strains from different clonal lineages in South Korea. Microb Drug Resist. 27:1271–81. DOI: 10.1089/mdr.2020.0346. PMID: 33691494.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Vancomycin Resistance of Staphylococcus aureus in Korean Primary Hospitals
- Vancomycin-resistant Staphylococcus aureus
- Synergy of Arbekacin-based Combinations Against Vancomycin Hetero-intermediate Staphylococcus aureus
- Molecular Typing and Resistance Profiles of Vancomycin-Intermediate Staphylococcus aureus in Korea: Results from a National Surveillance Study, 2007-2013
- Antimicrobial Resistance in Gram-positive Cocci: Past 50 Years, Present and Future