Obstet Gynecol Sci.
2023 Sep;66(5):417-429. 10.5468/ogs.22315.
Transcriptomic patterns in early-secretory and mid-secretory endometrium in a natural menstrual cycle immediately before in vitro fertilization and embryo transfer
- Affiliations
-
- 1Department of Obstetrics and Gynecology, Inha University Hospital, College of Medicine, Inha University, Incheon, Korea
- 2Department of Obstetrics and Gynecology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Obstetrics and Gynecology, Institute of Women’s Life Medical Science, Yonsei University College of Medicine, Seoul, Korea
- 4Department of Obstetrics and Gynecology, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 5Department of Obstetrics and Gynecology, Yongin Severance Hospital, Yonsei University College of Medicine, Yongin, Korea
- KMID: 2545888
- DOI: http://doi.org/10.5468/ogs.22315
Abstract
Objective
This study aimed to evaluate the endometrial transcriptomic patterns in the early secretory phase (ESP) and mid-secretory phase (MSP) of the natural menstrual cycle before in vitro fertilization and embryo transfer (IVF-ET).
Methods
Thirty patients whose endometrial tissues were obtained from the ESP or MSP of a natural menstrual cycle immediately before IVF-ET were included. Endometrial dating was histologically confirmed as ESP (cycle days 16-18) or MSP (cycle days 19-21), according to the noyes criteria. The patients were divided into two groups depending on the IVF-ET outcome: pregnant (n=14; 7 in ESP and 7 in MSP) or non-pregnant (n=16; 8 in ESP and 8 in MSP). Differentially expressed genes (DEGs) in the MSP, compared to the ESP, were identified using NanoString nCounter (NanoString Technologies, Seattle, WA, USA) data for both the pregnant and non-pregnant groups.
Results
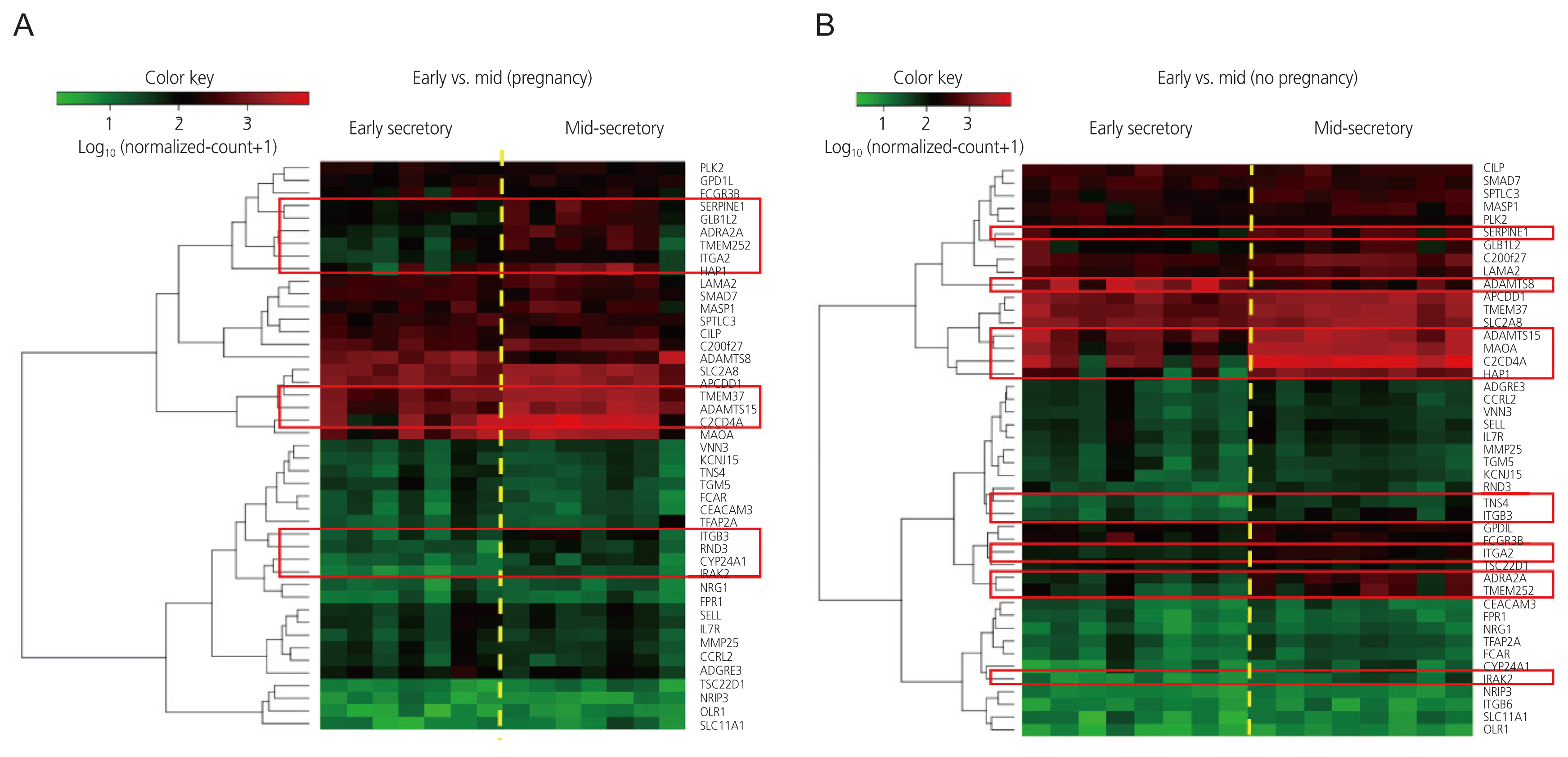
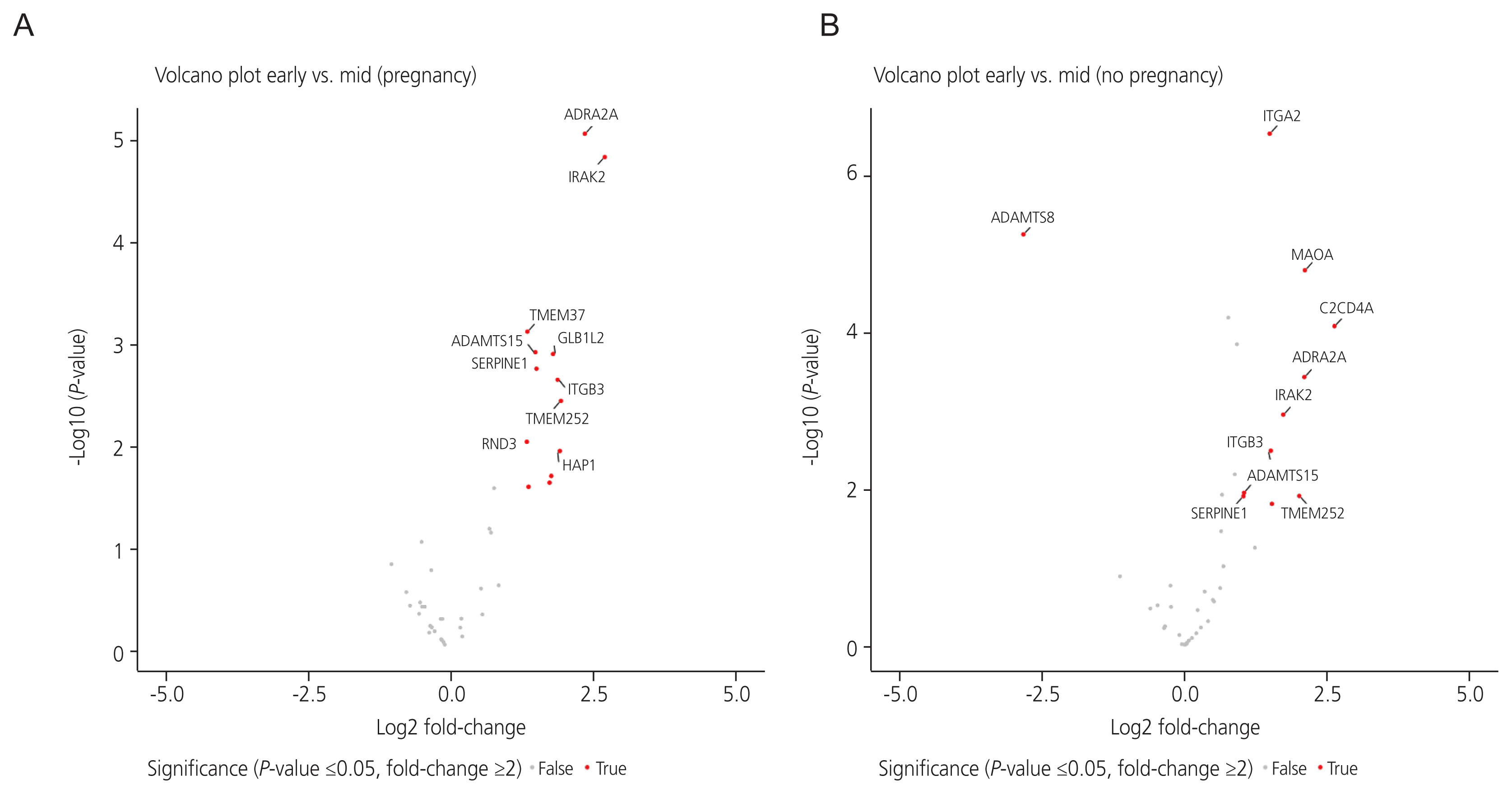
Thirteen DEGs in the pregnant group and 11 DEGs in the non-pregnant group were identified in the MSP compared to those in the ESP. In both groups, adrenoceptor alpha 2A, interleukin 1 receptor-associated kinase 2, a disintegrin and metalloproteinase with thrombospondin repeats 15 (ADAMTS15), serpin family E member 1, integrin subunit beta 3, transmembrane protein 252 (TMEM252), huntingtin associated protein 1, C2 calcium-dependent domain containing 4A, and integrin subunit alpha 2 were upregulated in the MSP, compared to the ESP. TMEM37, galactosidase beta 1 like 2, Rho family GTPase 3, and cytochrome P450 family 24 subfamily A member 1 were upregulated in the MSP only in the pregnant group. ADAMTS8 was downregulated and monoamine oxidase A was upregulated in the MSP only in the non-pregnant group.
Conclusion
Transcriptomic patterns in the endometrium immediately before IVF-ET appear to differ according to the IVF-ET outcome. These novel DEGs, which have not been previously studied, may have functional significance during the window of implantation and serve as potential biomarkers of endometrial receptivity.
Keyword
Figure
Reference
-
References
1. Norwitz ER, Schust DJ, Fisher SJ. Implantation and the survival of early pregnancy. N Engl J Med. 2001; 345:1400–8.2. Wilcox AJ, Baird DD, Weinberg CR. Time of implantation of the conceptus and loss of pregnancy. N Engl J Med. 1999; 340:1796–9.3. Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Am J Obstet Gynecol. 1975; 122:262–3.4. Talbi S, Hamilton AE, Vo KC, Tulac S, Overgaard MT, Dosiou C, et al. Molecular phenotyping of human endometrium distinguishes menstrual cycle phases and underlying biological processes in normo-ovulatory women. Endocrinology. 2006; 147:1097–121.5. Edwards RG. Human uterine endocrinology and the implantation window. Ann N Y Acad Sci. 1988; 541:445–54.6. Harper MJ. The implantation window. Baillieres Clin Obstet Gynaecol. 1992; 6:351–71.7. Coutifaris C, Myers ER, Guzick DS, Diamond MP, Carson SA, Legro RS, et al. Histological dating of timed endometrial biopsy tissue is not related to fertility status. Fertil Steril. 2004; 82:1264–72.8. Haouzi D, Mahmoud K, Fourar M, Bendhaou K, Dechaud H, De Vos J, et al. Identification of new biomarkers of human endometrial receptivity in the natural cycle. Hum Reprod. 2009; 24:198–205.9. Achache H, Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update. 2006; 12:731–46.10. Wang W, Vilella F, Alama P, Moreno I, Mignardi M, Isakova A, et al. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat Med. 2020; 26:1644–53.11. Craciunas L, Gallos I, Chu J, Bourne T, Quenby S, Brosens JJ, et al. Conventional and modern markers of endometrial receptivity: a systematic review and meta-analysis. Hum Reprod Update. 2019; 25:202–23.12. Díaz-Gimeno P, Ruiz-Alonso M, Blesa D, Bosch N, Martínez-Conejero JA, Alamá P, et al. The accuracy and reproducibility of the endometrial receptivity array is superior to histology as a diagnostic method for endometrial receptivity. Fertil Steril. 2013; 99:508–17.13. Díaz-Gimeno P, Horcajadas JA, Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A, et al. A genomic diagnostic tool for human endometrial receptivity based on the transcriptomic signature. Fertil Steril. 2011; 95:50–60e1. –15.14. Patel JA, Patel AJ, Banker JM, Shah SI, Banker MR. Personalized embryo transfer helps in improving in vitro fertilization/ICSI outcomes in patients with recurrent implantation failure. J Hum Reprod Sci. 2019; 12:59–66.15. Tan J, Kan A, Hitkari J, Taylor B, Tallon N, Warraich G, et al. The role of the endometrial receptivity array (ERA) in patients who have failed euploid embryo transfers. J Assist Reprod Genet. 2018; 35:683–92.16. Cozzolino M, Diaz-Gimeno P, Pellicer A, Garrido N. Evaluation of the endometrial receptivity assay and the preimplantation genetic test for aneuploidy in overcoming recurrent implantation failure. J Assist Reprod Genet. 2020; 37:2989–97.17. Eisman LE, Pisarska MD, Wertheimer S, Chan JL, Akopians AL, Surrey MW, et al. Clinical utility of the endometrial receptivity analysis in women with prior failed transfers. J Assist Reprod Genet. 2021; 38:645–50.18. Saxtorph MH, Hallager T, Persson G, Petersen KB, Eriksen JO, Larsen LG, et al. Assessing endometrial receptivity after recurrent implantation failure: a prospective controlled cohort study. Reprod Biomed Online. 2020; 41:998–1006.19. Simón C, Gómez C, Cabanillas S, Vladimirov I, Castillón G, Giles J, et al. A 5-year multicentre randomized controlled trial comparing personalized, frozen and fresh blastocyst transfer in IVF. Reprod Biomed Online. 2020; 41:402–15.20. Raff M, Jacobs E, Voorhis BV. End of an endometrial receptivity array? Fertil Steril. 2022; 118:737.21. Sebastian-Leon P, Garrido N, Remohí J, Pellicer A, Diaz-Gimeno P. Asynchronous and pathological windows of implantation: two causes of recurrent implantation failure. Hum Reprod. 2018; 33:626–35.22. Mirkin S, Arslan M, Churikov D, Corica A, Diaz JI, Williams S, et al. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum Reprod. 2005; 20:2104–17.23. Hu S, Yao G, Wang Y, Xu H, Ji X, He Y, et al. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J Clin Endocrinol Metab. 2014; 99:E2744–53.24. R Riesewijk A, Martín J, van Os R, Horcajadas JA, Polman J, Pellicer A, et al. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol Hum Reprod. 2003; 9:253–64.25. Carson DD, Lagow E, Thathiah A, Al-Shami R, Farach-Carson MC, Vernon M, et al. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol Hum Reprod. 2002; 8:871–9.26. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3:RESEARCH0034.27. Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016; 44:W83–9.28. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–7.29. Solimena M, Schulte AM, Marselli L, Ehehalt F, Richter D, Kleeberg M, et al. Systems biology of the IMIDIA biobank from organ donors and pancreatectomised patients defines a novel transcriptomic signature of islets from individuals with type 2 diabetes. Diabetologia. 2018; 61:641–57.30. Li C, Shen Z, Zhou Y, Yu W. Independent prognostic genes and mechanism investigation for colon cancer. Biol Res. 2018; 51:10.31. Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011; 12:449–62.32. Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, Chardin P. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol. 1998; 141:187–97.33. Liu B, Lin X, Yang X, Dong H, Yue X, Andrade KC, et al. Downregulation of RND3/RhoE in glioblastoma patients promotes tumorigenesis through augmentation of notch transcriptional complex activity. Cancer Med. 2015; 4:1404–16.34. Dai Y, Song J, Li W, Yang T, Yue X, Lin X, et al. RhoE fine-tunes inflammatory response in myocardial infarction. Circulation. 2019; 139:1185–98.35. Huang L, Ma Y, Chen L, Chang J, Zhong M, Wang Z, et al. Maternal RND3/RhoE deficiency impairs placental mitochondrial function in preeclampsia by modulating the PPARγ-UCP2 cascade. FASEB J. 2021; 35:e21555.36. Lin X, Liu B, Yang X, Yue X, Diao L, Wang J, et al. Genetic deletion of Rnd3 results in aqueductal stenosis leading to hydrocephalus through up-regulation of Notch signaling. Proc Natl Acad Sci U S A. 2013; 110:8236–41.37. Lartey J, Gampel A, Pawade J, Mellor H, Bernal AL. Expression of RND proteins in human myometrium. Biol Reprod. 2006; 75:452–61.38. Ji JL, Muyayalo KP, Zhang YH, Hu XH, Liao AH. Immunological function of vitamin D during human pregnancy. Am J Reprod Immunol. 2017; 78:e12716.39. Agic A, Xu H, Altgassen C, Noack F, Wolfler MM, Diedrich K, et al. Relative expression of 1,25-dihydroxyvitamin D3 receptor, vitamin D 1 alpha-hydroxylase, vitamin D 24-hydroxylase, and vitamin D 25-hydroxylase in endometriosis and gynecologic cancers. Reprod Sci. 2007; 14:486–97.40. Viganò P, Lattuada D, Mangioni S, Ermellino L, Vignali M, Caporizzo E, et al. Cycling and early pregnant endometrium as a site of regulated expression of the vitamin D system. J Mol Endocrinol. 2006; 36:415–24.41. Lan NC, Heinzmann C, Gal A, Klisak I, Orth U, Lai E, et al. Human monoamine oxidase A and B genes map to Xp 11.23 and are deleted in a patient with Norrie disease. Genomics. 1989; 4:552–9.42. Wu JB, Yin L, Shi C, Li Q, Duan P, Huang JM, et al. MAOA-dependent activation of Shh-IL6-RANKL signaling network promotes prostate cancer metastasis by engaging tumor-stromal cell interactions. Cancer Cell. 2017; 31:368–82.43. Henriquez S, Tapia A, Quezada M, Vargas M, Cardenas H, Rios M, et al. Deficient expression of monoamine oxidase A in the endometrium is associated with implantation failure in women participating as recipients in oocyte donation. Mol Hum Reprod. 2006; 12:749–54.44. Tian J, Zhang C, Kang N, Wang J, Kong N, Zhou J, et al. Attenuated monoamine oxidase a impairs endometrial receptivity in women with adenomyosis via downregulation of FOXO1. Biol Reprod. 2021; 105:1443–57.45. Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005; 386:15–27.46. Apte SS. A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motifs: the ADAMTS family. Int J Biochem Cell Biol. 2004; 36:981–5.47. Wagstaff L, Kelwick R, Decock J, Edwards DR. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front Biosci (Landmark Ed). 2011; 16:1861–72.48. Wen J, Zhu H, Leung PC. Gonadal steroids regulate the expression of aggrecanases in human endometrial stromal cells in vitro. J Cell Mol Med. 2013; 17:1325–34.49. Wu LL, Zhou XF. Huntingtin associated protein 1 and its functions. Cell Adh Migr. 2009; 3:71–6.50. Zhao X, Chen A, Wang Z, Xu XH, Tao Y. Biological functions and potential therapeutic applications of huntingtin-associated protein 1: progress and prospects. Clin Transl Oncol. 2022; 24:203–14.51. Li XJ, Li SH. HAP1 and intracellular trafficking. Trends Pharmacol Sci. 2005; 26:1–3.52. D’Aurora M, Romani F, Franchi S, Diomede F, Merciaro I, Impicciatore GG, et al. MRAP2 regulates endometrial receptivity and function. Gene. 2019; 703:7–12.53. Liu C, Li Y, Jiang H, Liu Y, Song X. The clinical outcomes of fresh versus frozen embryos transfer in women ≥40 years with poor ovarian response. Obstet Gynecol Sci. 2021; 64:284–92.54. Liu Y, Lee KF, Ng EH, Yeung WS, Ho PC. Gene expression profiling of human peri-implantation endometria between natural and stimulated cycles. Fertil Steril. 2008; 90:2152–64.55. Horcajadas JA, Riesewijk A, Polman J, van Os R, Pellicer A, Mosselman S, et al. Effect of controlled ovarian hyperstimulation in IVF on endometrial gene expression profiles. Mol Hum Reprod. 2005; 11:195–205.56. Horcajadas JA, Minguez P, Dopazo J, Esteban FJ, Dominguez F, Giudice LC, et al. Controlled ovarian stimulation induces a functional genomic delay of the endometrium with potential clinical implications. J Clin Endocrinol Metab. 2008; 93:4500–10.57. Haouzi D, Assou S, Mahmoud K, Tondeur S, Reme T, Hedon B, et al. Gene expression profile of human endometrial receptivity: comparison between natural and stimulated cycles for the same patients. Hum Reprod. 2009; 24:1436–45.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression pettern of Sex Hormone Receptors , Integrins , Cyclooxygenases ( COX ) in Human Endometrium During Menstrual Cycle
- Transvaginal Ultrasonographic Analysis of Endometrial Pattern and Thickness Changes in Normal Menstrual Cycle
- Expression of Nitric Oxide Synthase in the Endometrium During the Menstrual Cycle
- Cyclic Expression of Cyclooxygenase-1 and -2 in Human Endometrium
- Expression of Transforming Growth Factor-beta1 , beta2 by Immunohistochemical Staining method: In Human Endometrium through the Menstrual Cycle