Anat Cell Biol.
2023 Mar;56(1):54-60. 10.5115/acb.22.178.
Revealing Joseon period People’s single nucleotide polymorphism associated with lactase gene by ancient DNA analysis of human remains from archaeological sites in Korea
- Affiliations
-
- 1Department of Mortuary Science, College of Bio-Convergence, Eulji University, Seongnam, Korea
- 2Department of Anatomy, Dankook University College of Medicine, Cheonan, Korea
- 3Catholic Institute for Applied Anatomy, Department of Anatomy, Colllege of Medicine, The Catholic University of Korea, Seoul, Korea
- 4Nuri Institute of Archaeology, Gongju, Korea
- 5Sudo Institute of Cultural Heritage, Seoul, Korea
- 6Department of Forensic Medicine, Seoul National University College of Medicine, Seoul, Korea
- 7Institute of Forensic and Anthropological Science, Seoul National University College of Medicine, Seoul, Korea
- 8Department of Anatomy and Cell Biology, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2540985
- DOI: http://doi.org/10.5115/acb.22.178
Abstract
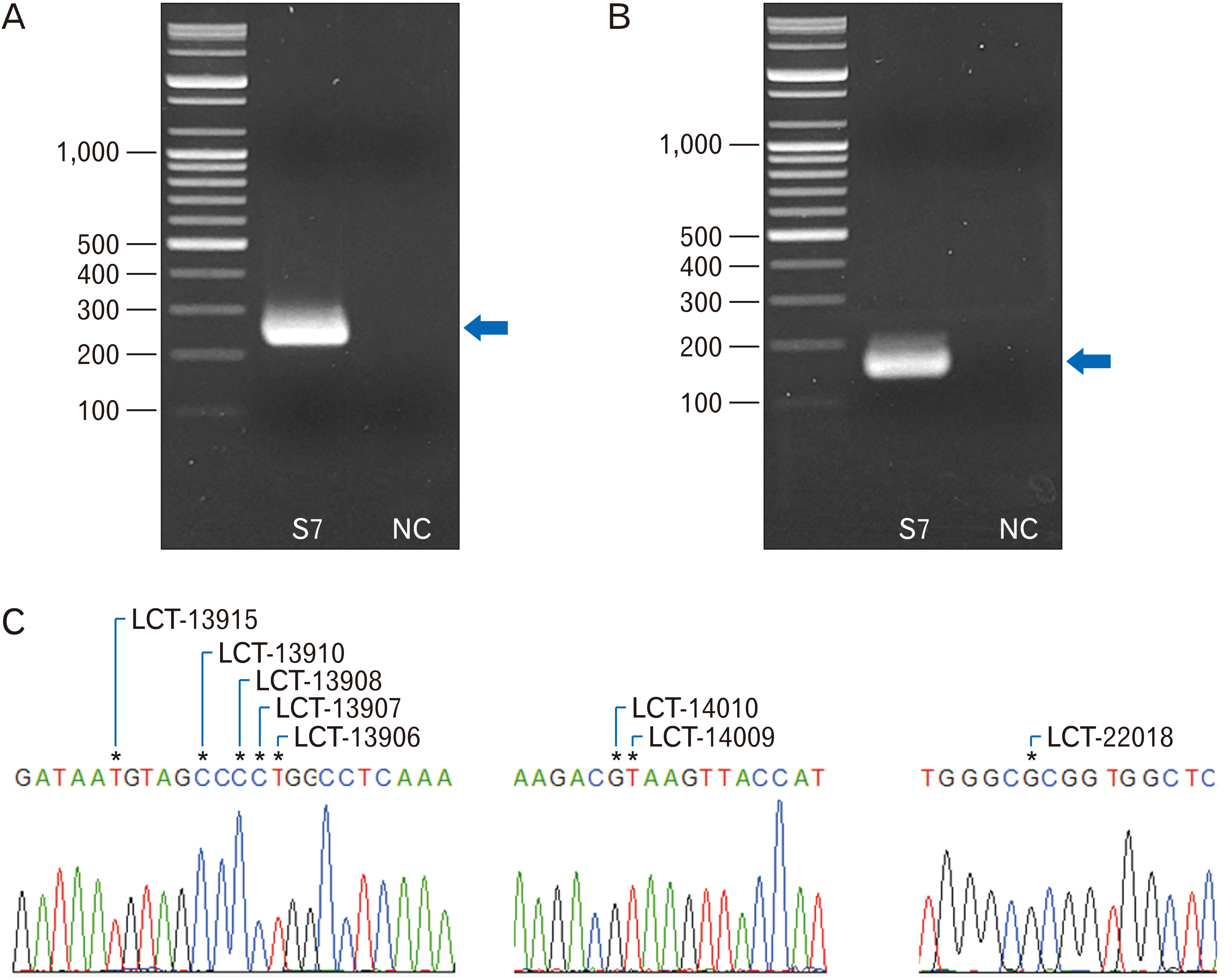
- Lactase non-persistence (LNP), one of the causes of lactose intolerance, is related to lactase gene associated single nucleotide polymorphisms (SNPs). Since the frequency of LNP varies by ethnic group and country, the research to reveal the presence or absence of LNP for specific people has been conducted worldwide. However, in East Asia, the study of lactase gene associated SNPs have not been sufficiently examined so far using ancient human specimens from archaeological sites. In our study of Joseon period human remains (n=14), we successfully revealed genetic information of lactase gene associated SNPs (rs1679771596, rs41525747, rs4988236, rs4988235, rs41380347, rs869051967, rs145946881 and rs182549), further confirming that as for eight SNPs, the pre-modern Korean people had a lactase non-persistent genotype. Our report contributes to the establishment of LNP associated SNP analysis technique that can be useful in forthcoming studies on human bones and mummy samples from East Asian archaeological sites.
Keyword
Figure
Reference
-
References
1. Lomer MC, Parkes GC, Sanderson JD. 2008; Review article: lactose intolerance in clinical practice--myths and realities. Aliment Pharmacol Ther. 27:93–103. DOI: 10.1111/j.1365-2036.2007.03557.x. PMID: 17956597.
Article2. Levitt M, Wilt T, Shaukat A. 2013; Clinical implications of lactose malabsorption versus lactose intolerance. J Clin Gastroenterol. 47:471–80. DOI: 10.1097/MCG.0b013e3182889f0f. PMID: 23632346.
Article3. Jarrett EC, Holman GH. 1966; Lactose absorption in the premature infant. Arch Dis Child. 41:525–7. DOI: 10.1136/adc.41.219.525. PMID: 5957726. PMCID: PMC2019603.
Article4. Bayless TM, Rosensweig NS. 1966; A racial difference in incidence of lactase deficiency. A survey of milk intolerance and lactase deficiency in healthy adult males. JAMA. 197:968–72. DOI: 10.1001/jama.1966.03110120074017. PMID: 5953213.
Article5. Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Järvelä I. 2002; Identification of a variant associated with adult-type hypolactasia. Nat Genet. 30:233–7. DOI: 10.1038/ng826. PMID: 11788828.
Article6. Järvelä IE. 2005; Molecular diagnosis of adult-type hypolactasia (lactase non-persistence). Scand J Clin Lab Invest. 65:535–9. DOI: 10.1080/00365510500208316. PMID: 16271984.
Article7. Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. 2009; Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 124:579–91. DOI: 10.1007/s00439-008-0593-6. PMID: 19034520.
Article8. Swallow DM. 2003; Genetics of lactase persistence and lactose intolerance. Annu Rev Genet. 37:197–219. DOI: 10.1146/annurev.genet.37.110801.143820. PMID: 14616060.
Article9. Raz M, Sharon Y, Yerushalmi B, Birk R. 2013; Frequency of LCT-13910C/T and LCT-22018G/A single nucleotide polymorphisms associated with adult-type hypolactasia/lactase persistence among Israelis of different ethnic groups. Gene. 519:67–70. DOI: 10.1016/j.gene.2013.01.049. PMID: 23415628.
Article10. Enattah NS, Trudeau A, Pimenoff V, Maiuri L, Auricchio S, Greco L, Rossi M, Lentze M, Seo JK, Rahgozar S, Khalil I, Alifrangis M, Natah S, Groop L, Shaat N, Kozlov A, Verschubskaya G, Comas D, Bulayeva K, Mehdi SQ, Terwilliger JD, Sahi T, Savilahti E, Perola M, Sajantila A, Järvelä I, Peltonen L. 2007; Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet. 81:615–25. DOI: 10.1086/520705. PMID: 17701907. PMCID: PMC1950831.
Article11. Sverrisdóttir OÓ, Timpson A, Toombs J, Lecoeur C, Froguel P, Carretero JM, Arsuaga Ferreras JL, Götherström A, Thomas MG. 2014; Direct estimates of natural selection in Iberia indicate calcium absorption was not the only driver of lactase persistence in Europe. Mol Biol Evol. 31:975–83. DOI: 10.1093/molbev/msu049. PMID: 24448642.
Article12. Allentoft ME, Sikora M, Sjögren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas AS, Margaryan A, Higham T, Chivall D, Lynnerup N, Harvig L, Baron J, Della Casa P, Dąbrowski P, Duffy PR, Ebel AV, Epimakhov A, Frei K, Furmanek M, Gralak T, Gromov A, Gronkiewicz S, Grupe G, Hajdu T, Jarysz R, Khartanovich V, Khokhlov A, Kiss V, Kolář J, Kriiska A, Lasak I, Longhi C, McGlynn G, Merkevicius A, Merkyte I, Metspalu M, Mkrtchyan R, Moiseyev V, Paja L, Pálfi G, Pokutta D, Pospieszny Ł, Price TD, Saag L, Sablin M, Shishlina N, Smrčka V, Soenov VI, Szeverényi V, Tóth G, Trifanova SV, Varul L, Vicze M, Yepiskoposyan L, Zhitenev V, Orlando L, Sicheritz-Pontén T, Brunak S, Nielsen R, Kristiansen K, Willerslev E. 2015; Population genomics of Bronze Age Eurasia. Nature. 522:167–72. DOI: 10.1038/nature14507. PMID: 26062507.
Article13. Jeong C, Wilkin S, Amgalantugs T, Bouwman AS, Taylor WTT, Hagan RW, Bromage S, Tsolmon S, Trachsel C, Grossmann J, Littleton J, Makarewicz CA, Krigbaum J, Burri M, Scott A, Davaasambuu G, Wright J, Irmer F, Myagmar E, Boivin N, Robbeets M, Rühli FJ, Krause J, Frohlich B, Hendy J, Warinner C. 2018; Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe. Proc Natl Acad Sci U S A. 115:E11248–55. DOI: 10.1073/pnas.1813608115. PMID: 30397125. PMCID: PMC6275519.
Article14. Saag L, Laneman M, Varul L, Malve M, Valk H, Razzak MA, Shirobokov IG, Khartanovich VI, Mikhaylova ER, Kushniarevich A, Scheib CL, Solnik A, Reisberg T, Parik J, Saag L, Metspalu E, Rootsi S, Montinaro F, Remm M, Mägi R, D'Atanasio E, Crema ER, Díez-Del-Molino D, Thomas MG, Kriiska A, Kivisild T, Villems R, Lang V, Metspalu M, Tambets K. 2019; The arrival of Siberian ancestry connecting the eastern Baltic to Uralic speakers further east. Curr Biol. 29:1701–11.e16. DOI: 10.1016/j.cub.2019.04.026. PMID: 31080083. PMCID: PMC6544527.
Article15. Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. 2001; Ancient DNA. Nat Rev Genet. 2:353–9. DOI: 10.1038/35072071. PMID: 11331901.
Article16. Willerslev E, Cooper A. 2005; Ancient DNA. Proc Biol Sci. 272:3–16. DOI: 10.1098/rspb.2004.2813. PMID: 15875564. PMCID: PMC1634942.17. Kim YS, Oh CS, Lee SJ, Park JB, Kim MJ, Shin DH. 2011; Sex determination of Joseon people skeletons based on anatomical, cultural and molecular biological clues. Ann Anat. 193:539–43. DOI: 10.1016/j.aanat.2011.07.002. PMID: 21889322.
Article18. Oh CS, Hong JH, Shin DH. 2019; Mitochondrial DNA analysis of the human skeletons from Goryeo dynasty graves discovered at Youngwol, Gangwon-do. Anat Biol Anthropol. 32:61–7. DOI: 10.11637/aba.2019.32.2.61.
Article19. Kato K, Ishida S, Tanaka M, Mitsuyama E, Xiao JZ, Odamaki T. 2018; Association between functional lactase variants and a high abundance of Bifidobacterium in the gut of healthy Japanese people. PLoS One. 13:e0206189. DOI: 10.1371/journal.pone.0206189. PMID: 30339693. PMCID: PMC6195297.
Article20. Peng MS, He JD, Zhu CL, Wu SF, Jin JQ, Zhang YP. 2012; Lactase persistence may have an independent origin in Tibetan populations from Tibet, China. J Hum Genet. 57:394–7. DOI: 10.1038/jhg.2012.41. PMID: 22572735.
Article21. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018; MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–9. DOI: 10.1093/molbev/msy096. PMID: 29722887. PMCID: PMC5967553.
Article22. Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. 2007; Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci U S A. 104:3736–41. DOI: 10.1073/pnas.0607187104. PMID: 17360422. PMCID: PMC1820653.
Article23. Nagy D, Tömöry G, Csányi B, Bogácsi-Szabó E, Czibula Á, Priskin K, Bede O, Bartosiewicz L, Downes CS, Raskó I. 2011; Comparison of lactase persistence polymorphism in ancient and present-day Hungarian populations. Am J Phys Anthropol. 145:262–9. DOI: 10.1002/ajpa.21490. PMID: 21365615.
Article24. Plantinga TS, Alonso S, Izagirre N, Hervella M, Fregel R, van der Meer JW, Netea MG, de la Rúa C. 2012; Low prevalence of lactase persistence in Neolithic South-West Europe. Eur J Hum Genet. 20:778–82. Erratum in: Eur J Hum Genet 2012;20:810. DOI: 10.1038/ejhg.2011.254. PMID: 22234158. PMCID: PMC3376259.
Article25. Krüttli A, Bouwman A, Akgül G, Della Casa P, Rühli F, Warinner C. 2014; Ancient DNA analysis reveals high frequency of European lactase persistence allele (T-13910) in medieval central europe. PLoS One. 9:e86251. DOI: 10.1371/journal.pone.0086251. PMID: 24465990. PMCID: PMC3900515. PMID: d475bd5f2ed640f3b8ccbe524cd52e65.
Article26. Płoszaj T, Jędrychowska-Dańska K, Zamerska A, Drozd-Lipińska A, Poliński D, Janowski A, Witas H. 2017; Ancient DNA analysis might suggest external origin of individuals from chamber graves placed in medieval cemetery in Pień, Central Poland. Anthropol Anz. 74:319–37. DOI: 10.1127/anthranz/2017/0727. PMID: 28799621.
Article27. Mnich B, Spinek AE, Chyleński M, Sommerfeld A, Dabert M, Juras A, Szostek K. 2018; Analysis of LCT-13910 genotypes and bone mineral density in ancient skeletal materials. PLoS One. 13:e0194966. Erratum in: PLoS One 2020;15:e0236908. DOI: 10.1371/journal.pone.0236908. PMID: 32702066. PMCID: PMC7377453. PMID: c071ffd7bba3433c8f2f9e7f45053b97.
Article28. Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, Leidinger P, Backes C, Khairat R, Forster M, Stade B, Franke A, Mayer J, Spangler J, McLaughlin S, Shah M, Lee C, Harkins TT, Sartori A, Moreno-Estrada A, Henn B, Sikora M, Semino O, Chiaroni J, Rootsi S, Myres NM, Cabrera VM, Underhill PA, Bustamante CD, Vigl EE, Samadelli M, Cipollini G, Haas J, Katus H, O'Connor BD, Carlson MR, Meder B, Blin N, Meese E, Pusch CM, Zink A. 2012; New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 3:698. DOI: 10.1038/ncomms1701. PMID: 22426219.
Article29. Jeong C, Wang K, Wilkin S, Taylor WTT, Miller BK, Bemmann JH, Stahl R, Chiovelli C, Knolle F, Ulziibayar S, Khatanbaatar D, Erdenebaatar D, Erdenebat U, Ochir A, Ankhsanaa G, Vanchigdash C, Ochir B, Munkhbayar C, Tumen D, Kovalev A, Kradin N, Bazarov BA, Miyagashev DA, Konovalov PB, Zhambaltarova E, Miller AV, Haak W, Schiffels S, Krause J, Boivin N, Erdene M, Hendy J, Warinner C. 2020; A dynamic 6,000-year genetic history of Eurasia's Eastern Steppe. Cell. 183:890–904.e29. DOI: 10.1016/j.cell.2020.10.015. PMID: 33157037. PMCID: PMC7664836.
Article30. Ning C, Li T, Wang K, Zhang F, Li T, Wu X, Gao S, Zhang Q, Zhang H, Hudson MJ, Dong G, Wu S, Fang Y, Liu C, Feng C, Li W, Han T, Li R, Wei J, Zhu Y, Zhou Y, Wang CC, Fan S, Xiong Z, Sun Z, Ye M, Sun L, Wu X, Liang F, Cao Y, Wei X, Zhu H, Zhou H, Krause J, Robbeets M, Jeong C, Cui Y. 2020; Ancient genomes from northern China suggest links between subsistence changes and human migration. Nat Commun. 11:2700. DOI: 10.1038/s41467-020-16557-2. PMID: 32483115. PMCID: PMC7264253. PMID: 952b238525f04d65a6fd937f71e31d06.
Article31. Holland MM, Huffine EF. 2001; Molecular analysis of the human mitochondrial DNA control region for forensic identity testing. Curr Protoc Hum Genet. Chapter 14:Unit 14.7. DOI: 10.1002/0471142905.hg1407s74. PMID: 22786611.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Bioarchaeological Studies on Ancient Human Skeletons in Korea
- Forensic Archeological Analysis on Human Remains from the Peculiar Burial Situation of Archaeological Excavation Site in Joseon Dynasty
- Mitochondrial DNA Analysis of the Human Skeletons from Goryeo Dynasty Graves Discovered at Youngwol, Gangwon-do
- Animal Bones Found at Gongpyeong-dong Archaeological Site, the Capital Area of Joseon Dynasty Period
- Forensic Anthropological Study on Saw Marks Appearing on the Tibiae of a Joseon Skeleton