J Korean Med Sci.
2023 Mar;38(11):e77. 10.3346/jkms.2023.38.e77.
Identifying Disease of Interest With Deep Learning Using Diagnosis Code
- Affiliations
-
- 1Department of Artificial Intelligence, Chung-Ang University, Seoul, Korea
- 2Department of Data Science, Sejong University, Seoul, Korea
- 3Department of Medicine, University of Virginia, Charlottesville, VA, USA
- 4Graduate School of Data Science, Seoul National University, Seoul, Korea
- 5Department of Medicine, University of Washington, Seattle, WA, USA
- KMID: 2540649
- DOI: http://doi.org/10.3346/jkms.2023.38.e77
Abstract
- Background
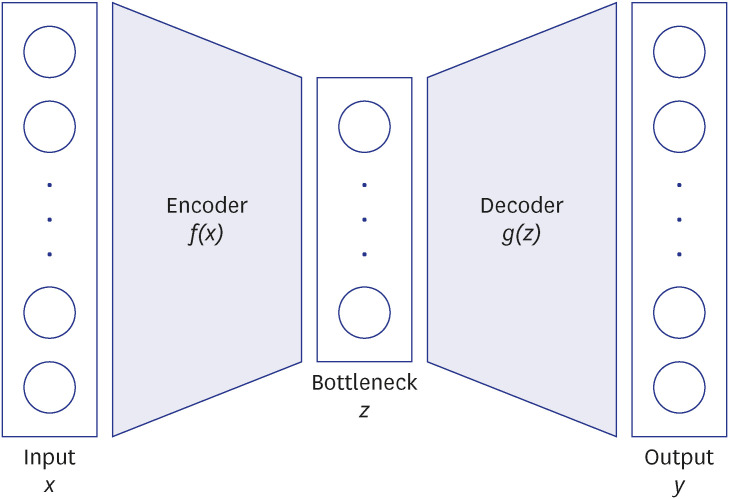
Autoencoder (AE) is one of the deep learning techniques that uses an artificial neural network to reconstruct its input data in the output layer. We constructed a novel supervised AE model and tested its performance in the prediction of a co-existence of the disease of interest only using diagnostic codes.
Methods
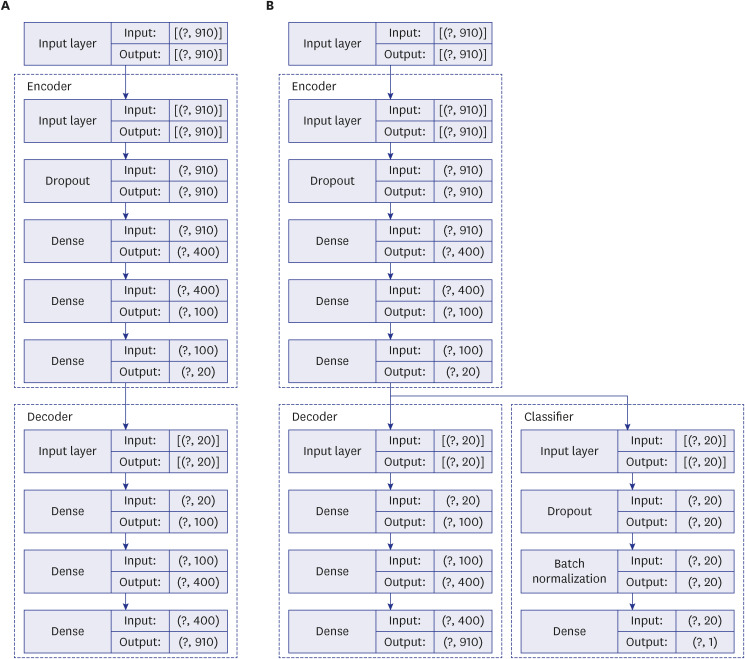
Diagnostic codes of one million randomly sampled patients listed in the Korean National Health Information Database in 2019 were used to train, validate, and test the prediction model. The first used AE solely for a feature engineering tool for an input of a classifier. Supervised Multi-Layer Perceptron (sMLP) was added to train a classifier to predict a binary level with latent representation as an input (AE + sMLP). The second model simultaneously updated the parameters in the AE and the connected MLP classifier during the learning process (End-to-End Supervised AE [EEsAE]). We tested the performances of these two models against baseline models, eXtreme Gradient Boosting (XGB) and naïve Bayes, in the prediction of co-existing gastric cancer diagnosis.
Results
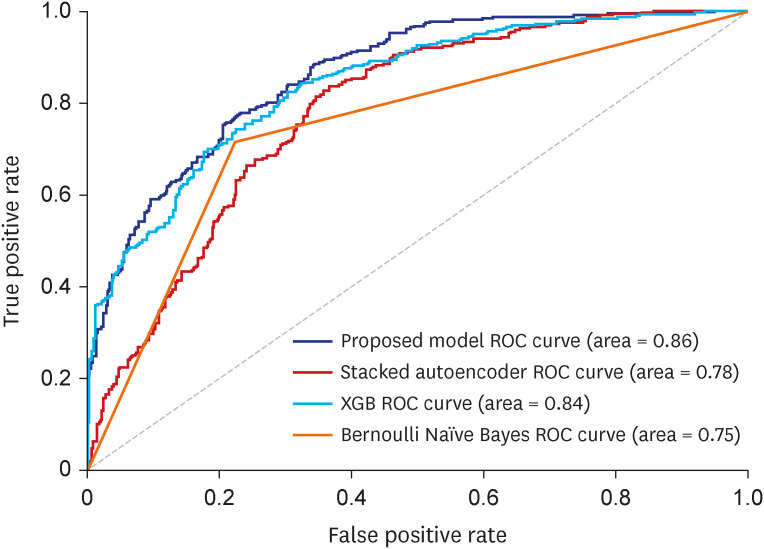
The proposed EEsAE model yielded the highest F1-score and highest area under the curve (0.86). The EEsAE and AE + sMLP gave the highest recalls. XGB yielded the highest precision. Ablation study revealed that iron deficiency anemia, gastroesophageal reflux disease, essential hypertension, gastric ulcers, benign prostate hyperplasia, and shoulder lesion were the top 6 most influential diagnoses on performance.
Conclusion
A novel EEsAE model showed promising performance in the prediction of a disease of interest.
Figure
Reference
-
1. Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019; 25(1):24–29. PMID: 30617335.2. Trabelsi A, Chaabane M, Ben-Hur A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics. 2019; 35(14):i269–i277. PMID: 31510640.3. Montesinos-López OA, Montesinos-López A, Pérez-Rodríguez P, Barrón-López JA, Martini JW, Fajardo-Flores SB, et al. A review of deep learning applications for genomic selection. BMC Genomics. 2021; 22(1):19. PMID: 33407114.4. Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019; 19(1):281. PMID: 31864346.5. Singhal A, Sinha P, Pant R. Use of deep learning in modern recommendation system: a summary of recent works. Int J Comput Appl. 2017; 108(7):17–22.6. Wang Z, Yu X, Feng N, Wang Z. An improved collaborative movie recommendation system using computational intelligence. J Vis Lang Comput. 2014; 25(6):667–675.7. Davidson J, Liebald B, Liu J, Nandy P, Van Vleet T, Gargi U, et al. The YouTube video recommendation system. In : Proceedings of the Fourth ACM Conference on Recommender; 2010 Sep 26–30; Barcelona, Spain. New York, NY, USA: ACM Press;2010. p. 293–296.8. Chen RC, Huang YH, Bau CT, Chen SM. A recommendation system based on domain ontology and SWRL for anti-diabetic drugs selection. Expert Syst Appl. 2012; 39(4):3995–4006.9. Doulaverakis C, Nikolaidis G, Kleontas A, Kompatsiaris I. Panacea, a semantic-enabled drug recommendations discovery framework. J Biomed Semantics. 2014; 5(1):13. PMID: 24602515.10. Shin A, Kim J, Park S. Gastric cancer epidemiology in Korea. J Gastric Cancer. 2011; 11(3):135–140. PMID: 22076217.11. Kweon SS. Updates on cancer epidemiology in Korea, 2018. Chonnam Med J. 2018; 54(2):90–100. PMID: 29854674.12. Shin DW, Cho B, Guallar E.. Korean National Health Insurance Database. JAMA Intern Med. 2016; 176(1):138.13. Lee J, Lee JS, Park SH, Shin SA, Kim K. Cohort profile: the National Health Insurance Service-National Sample Cohort (NHIS-NSC), South Korea. Int J Epidemiol. 2017; 46(2):e15. PMID: 26822938.14. Lee YS, Lee YR, Chae Y, Park SY, Oh IH, Jang BH. Translation of Korean medicine use to ICD-codes using National Health Insurance Service-National Sample Cohort. Evid Based Complement Alternat Med. 2016; 2016:8160838. PMID: 27069494.15. Simidjievski N, Bodnar C, Tariq I, Scherer P, Andres Terre H, Shams Z, et al. Variational autoencoders for cancer data integration: design principles and computational practice. Front Genet. 2019; 10:1205. PMID: 31921281.16. Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016; 6(1):26094. PMID: 27185194.17. Azimi-Sadjadi MR, Citrin S, Sheedvash S. Supervised learning process of multi-layer perceptron neural networks using fast least squares. Proc IEEE Int Conf Acoust Speech Signal Process. 1990; 3:1381–1384.18. Chen R, Song Y, Huang J, Wang J, Sun H, Wang H. Rapid diagnosis and continuous monitoring of intracerebral hemorrhage with magnetic induction tomography based on stacked autoencoder. Rev Sci Instrum. 2021; 92(8):084707. PMID: 34470442.19. Li D, Fu Z, Xu J. Stacked-autoencoder-based model for COVID-19 diagnosis on CT images. Appl Intell. 2021; 51(5):2805–2817.20. Quan H, Li B, Saunders LD, Parsons GA, Nilsson CI, Alibhai A, et al. Assessing validity of ICD-9-CM and ICD-10 administrative data in recording clinical conditions in a unique dually coded database. Health Serv Res. 2008; 43(4):1424–1441. PMID: 18756617.21. O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Serv Res. 2005; 40(5 Pt 2):1620–1639. PMID: 16178999.22. Tang GH, Hart R, Sholzberg M, Brezden-Masley C. Iron deficiency anemia in gastric cancer: a Canadian retrospective review. Eur J Gastroenterol Hepatol. 2018; 30(12):1497–1501. PMID: 30179903.23. Jeong O, Park YK, Ryu SY. Prevalence, severity, and evolution of postsurgical anemia after gastrectomy, and clinicopathological factors affecting its recovery. J Korean Surg Soc. 2012; 82(2):79–86. PMID: 22347709.24. Jung MJ, Kim HI, Cho HW, Yoon HY, Kim CB. Pre- and post-gastrectomy anemia in gastric cancer patients. Korean J Clin Oncol. 2011; 7(2):88–95.25. Kim JJ. Upper gastrointestinal cancer and reflux disease. J Gastric Cancer. 2013; 13(2):79–85. PMID: 23844321.26. Hansson LE. Risk of stomach cancer in patients with peptic ulcer disease. World J Surg. 2000; 24(3):315–320. PMID: 10658066.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- An Open Medical Platform to Share Source Code and Various Pre-Trained Weights for Models to Use in Deep Learning Research
- Developing and Evaluating Deep Learning Algorithms for Object Detection: Key Points for Achieving Superior Model Performance
- Current status of deep learning applications in abdominal ultrasonography
- Deep Learning in Dental Radiographic Imaging
- Deep Learning in MR Motion Correction:a Brief Review and a New Motion Simulation Tool (view2Dmotion)