J Liver Cancer.
2022 Sep;22(2):103-114. 10.17998/jlc.2022.09.08.
Liquid biopsy for early detection and therapeutic monitoring of hepatocellular carcinoma
- Affiliations
-
- 1LepiDyne Co., Ltd., Seoul, Korea
- 2Department of Biochemistry, Yonsei University, Seoul, Korea
- KMID: 2534237
- DOI: http://doi.org/10.17998/jlc.2022.09.08
Abstract
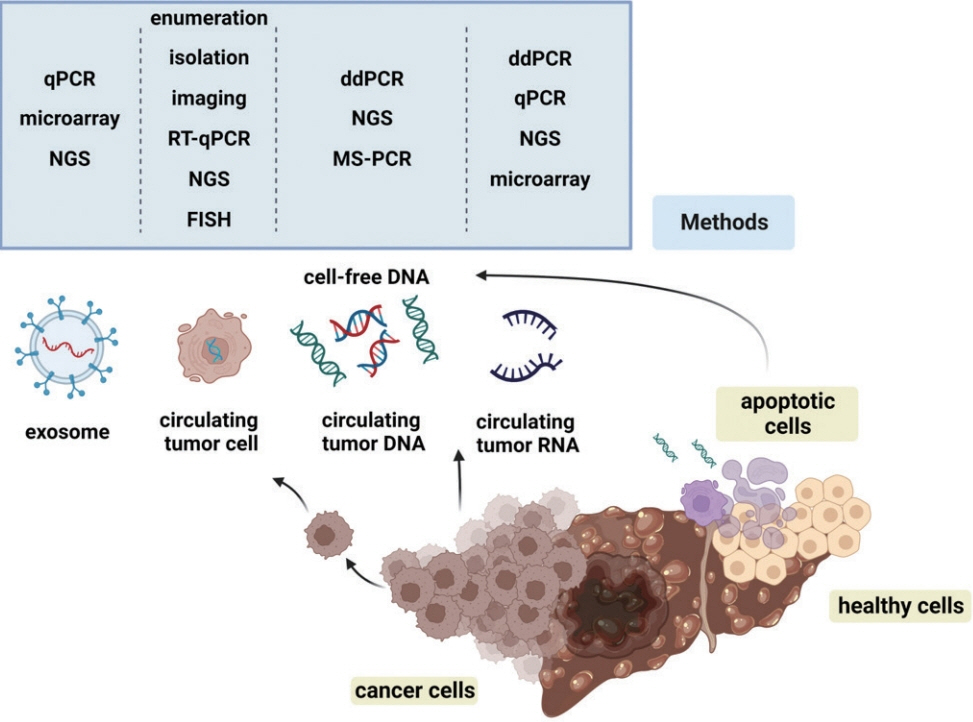
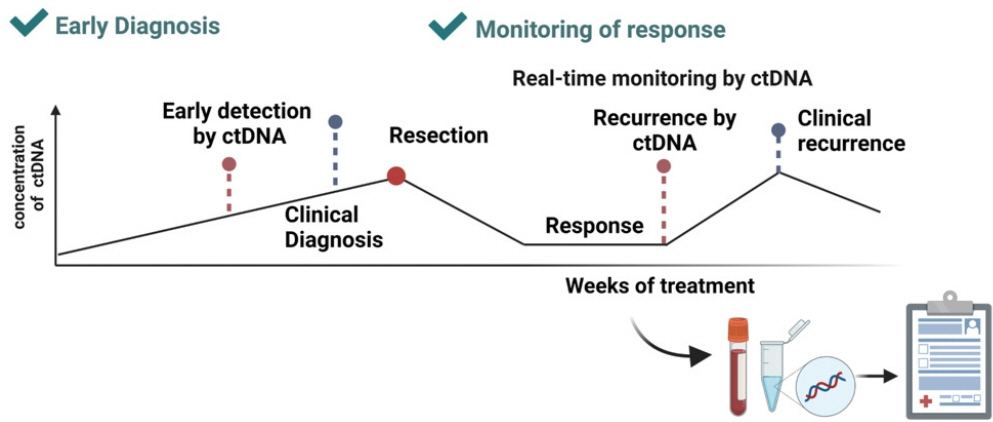
- Advances in our knowledge of the molecular characteristics of hepatocellular carcinoma (HCC) have enabled significant progress in the detection and therapeutic prediction of HCC. As a non-invasive alternative to tissue biopsy, liquid biopsy examines circulating cellular components such as exosomes, nucleic acids, and cell-free DNA found in body fluids (e.g., urine, saliva, ascites, and pleural effusions) and provides information about tumor characteristics. Technical advances in liquid biopsy have led to the increasing adoption of diagnostic and monitoring applications for HCC. This review summarizes the various analytes, ongoing clinical trials, and case studies of United States Food and Drug Administrationapproved in vitro diagnostic applications for liquid biopsy, and provides insight into its implementation in managing HCC.
Keyword
Figure
Cited by 1 articles
-
Current status of ultrasonography in national cancer surveillance program for hepatocellular carcinoma in South Korea: a large-scale multicenter study
Sun Hong Yoo, Soon Sun Kim, Sang Gyune Kim, Jung Hyun Kwon, Han-Ah Lee, Yeon Seok Seo, Young Kul Jung, Hyung Joon Yim, Do Seon Song, Seong Hee Kang, Moon Young Kim, Young-Hwan Ahn, Jieun Han, Young Seok Kim, Young Chang, Soung Won Jeong, Jae Young Jang, Jeong-Ju Yoo
J Liver Cancer. 2023;23(1):189-201. doi: 10.17998/jlc.2023.03.11.
Reference
-
1. International Agency for Research on Cancer. Estimated number of deaths in 2020, World, both sexes, all ages (excl. NMSC) [Internet]. Geneva (CH): World Health Organization;[cited 2022 Aug 1]. Available from: https://gco.iarc.fr/today/online-analysistable?v=2020&mode=cancer&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=39&type=1&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=0&ages_group%5B%5D=17&group_cancer=1&include_nmsc=0&include_nmsc_other=1.2. Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, et al. Annual report to the nation on the status of cancer, 1975-2014, featuring survival. J Natl Cancer Inst. 2017; 109:djx030.3. National Cancer Center. Annual report of cancer statistics in Korea in 2018 [Internet]. Goyang (KR): National Cancer Center;[cited 2022 Aug 1]. Available from: https://ncc.re.kr/cancerStatsView.ncc?bbsnum=558&searchKey=total&searchValue=&pageNum=1.4. Chon YE, Jeong SW, Jun DW. Hepatocellular carcinoma statistics in South Korea. Clin Mol Hepatol. 2021; 27:512–514.5. Statistics Korea. Preliminary results of birth and death statistics in 2021 [Internet]. Daejeon (KR): Statistics Korea;[cited 2022 Aug 1]. Available from: http://kostat.go.kr/portal/eng/pressReleases/8/10/index.board?bmode=read&bSeq=&aSeq=417980&pageNo=1&rowNum=10&navCount=10&currPg=&searchInfo=&sTarget=title&sTxt=.6. Cha C, Dematteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005; 19:25–37.7. Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981; 2:1129–1133.8. Niederau C, Lange S, Heintges T, Erhardt A, Buschkamp M, Hürter D, et al. Prognosis of chronic hepatitis C: results of a large, prospective cohort study. Hepatology. 1998; 28:1687–1695.9. Estes C, Razavi H, Loomba R, Younossi Z, Sanyal AJ. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology. 2018; 67:123–133.10. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Metaanalytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016; 64:73–84.11. Kanwal F, Kramer JR, Mapakshi S, Natarajan Y, Chayanupatkul M, Richardson PA, et al. Risk of hepatocellular cancer in patients with non-alcoholic fatty liver disease. Gastroenterology. 2018; 155:1828–1837.e2.12. Korean Association for the Study of the Liver. KASL clinical practice guidelines: management of chronic hepatitis B. Clin Mol Hepatol. 2016; 22:18–75.13. Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004; 130:417–422.14. Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a metaanalysis. Gastroenterology. 2018; 154:1706–1718. e1.15. Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017; 16:1155–1161.16. Macías M, Alegre E, Díaz-Lagares A, Patiño A, Pérez-Gracia JL, Sanmamed M, et al. Liquid biopsy: from basic research to clinical practice. Adv Clin Chem. 2018; 83:73–119.17. Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 2018; 109:2364–2374.18. Yang C, Xia BR, Jin WL, Lou G. Circulating tumor cells in precision oncology: clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 2019; 19:341.19. García-Pardo M, Makarem M, Li JJN, Kelly D, Leighl NB. Integrating circulating-free DNA (cfDNA) analysis into clinical practice: opportunities and challenges. Br J Cancer. 2022; 127:592–602.20. U.S. Food and Drug Administration. Companion diagnostics [Internet]. Silver Spring (MD): U.S. Food and Drug Administration;[cited 2022 Aug 1]. Available from: https://www.fda.gov/medicaldevices/in-vitro-diagnostics/companion-diagnostics.21. U.S. Food and Drug Administration. List of cleared or approved companion diagnostic devices (in vitro and imaging tools) [Internet]. Silver Spring (MD): U.S. Food and Drug Administration;[cited 2022 Aug 1]. Available from: https://www.fda.gov/medicaldevices/in-vitro-diagnostics/list-cleared-or-approved-companiondiagnostic-devices-in-vitro-and-imaging-tools.22. The ASCO Post. The evolution of liquid biopsy in cancer care [Internet]. Huntington (NY): The ASCO Post;[cited 2022 Sep 2]. Available from: https://ascopost.com/issues/october-10-2021/theevolution-of-liquid-biopsy-in-cancer-care/.23. Testing.com. Liquid biopsy [Internet]. Seattle (WA): Testing.com;[cited 2022 Aug 1]. Available from: https://www.testing.com/tests/liquid-biopsy/.24. Grabuschnig S, Bronkhorst AJ, Holdenrieder S, Rosales Rodriguez I, Schliep KP, Schwendenwein D, et al. Putative origins of cell-free DNA in humans: a review of active and passive nucleic acid release mechanisms. Int J Mol Sci. 2020; 21:8062.25. Mandel P, Metais P. Nuclear acids in human blood plasma. C R Seances Soc Biol Fil. 1948; 142:241–243.26. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977; 37:646–650.27. Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011; 11:426–437.28. Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016; 35:347–376.29. Alunni-Fabbroni M, Rönsch K, Huber T, Cyran CC, Seidensticker M, Mayerle J, et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: a translational exploratory study from the SORAMIC trial. J Transl Med. 2019; 17:328.30. van der Pol Y, Mouliere F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell. 2019; 36:350–368.31. Corcoran RB, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med. 2018; 379:1754–1765.32. Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017; 14:155–167.33. Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment length of circulating tumor DNA. PLoS Genet. 2016; 12:e1006162.34. Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018; 10:eaat4921.35. Lyu X, Tsui YM, Ho DW, Ng IO. Liquid biopsy using cell-free or circulating tumor DNA in the management of hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2022; 13:1611–1624.36. Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020; 9:1370.37. Nakamura Y, Shitara K. Development of circulating tumour DNA analysis for gastrointestinal cancers. ESMO Open. 2020; 5(Suppl 1):e000600.38. Koboldt DC. Best practices for variant calling in clinical sequencing. Genome Med. 2020; 12:91.39. Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer. 2021; 124:345–358.40. Ou CY, Vu T, Grunwald JT, Toledano M, Zimak J, Toosky M, et al. An ultrasensitive test for profiling circulating tumor DNA using integrated comprehensive droplet digital detection. Lab Chip. 2019; 19:993–1005.41. Gao Q, Zeng Q, Wang Z, Li C, Xu Y, Cui P, et al. Circulating cell-free DNA for cancer early detection. Innovation (Camb). 2022; 3:100259.42. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019; 570:385–389.43. Mathios D, Johansen JS, Cristiano S, Medina JE, Phallen J, Larsen KR, et al. Detection and characterization of lung cancer using cellfree DNA fragmentomes. Nat Commun. 2021; 12:5060.44. Lim HY, Heo J, Choi HJ, Lin CY, Yoon JH, Hsu C, et al. A phase II study of the efficacy and safety of the combination therapy of the MEK inhibitor refametinib (BAY 86-9766) plus sorafenib for Asian patients with unresectable hepatocellular carcinoma. Clin Cancer Res. 2014; 20:5976–5985.45. Li X, Xiang Y, Li F, Yin C, Li B, Ke X. WNT/β-catenin signaling pathway regulating T cell-inflammation in the tumor microenvironment. Front Immunol. 2019; 10:2293.46. Hlady RA, Zhou D, Puszyk W, Roberts LR, Liu C, Robertson KD. Initiation of aberrant DNA methylation patterns and heterogeneity in precancerous lesions of human hepatocellular cancer. Epigenetics. 2017; 12:215–225.47. Han LY, Fan YC, Mu NN, Gao S, Li F, Ji XF, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) is a potential biomarker for hepatitis B Virus associated hepatocellular carcinoma. Int J Med Sci. 2014; 11:164–171.48. Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards V DK, Roberts LR, et al. A novel blood-based panel of methylated DNA and protein markers for detection of early-stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2021; 19:2597–2605. e4.49. Chalasani NP, Porter K, Bhattacharya A, Book AJ, Neis BM, Xiong KM, et al. Validation of a novel multitarget blood test shows high sensitivity to detect early stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2022; 20:173–182. e7.50. Lin N, Lin Y, Xu J, Liu D, Li D, Meng H, et al. A multi-analyte cellfree DNA-based blood test for early detection of hepatocellular carcinoma. Hepatol Commun. 2022; 6:1753–1763.51. Rosen AW, Gögenur M, Paulsen IW, Olsen J, Eiholm S, Kirkeby LT, et al. Perioperative changes in cell-free DNA for patients undergoing surgery for colon cancer. BMC Gastroenterol. 2022; 22:168.52. Bolhuis K, van ‘t Erve I, Mijnals C, Delis-Van Diemen PM, Huiskens J, Komurcu A, et al. Postoperative circulating tumour DNA is associated with pathologic response and recurrence-free survival after resection of colorectal cancer liver metastases. EBioMedicine. 2021; 70:103498.53. Sefrioui D, Verdier V, Savoye-Collet C, Beaussire L, Ghomadi S, Gangloff A, et al. Circulating DNA changes are predictive of disease progression after transarterial chemoembolization. Int J Cancer. 2022; 150:532–541.54. Li Z, Xiao D, Li X, Zhan P, Wang J, Zhang H. Early recurrence detected in hepatocellular carcinoma patients after transcatheter arterial chemoembolization treatment with plasma cell-free DNA. Eur J Gastroenterol Hepatol. 2019; 31:885–892.55. Clouthier DL, Lien SC, Yang SYC, Nguyen LT, Manem VSK, Gray D, et al. An interim report on the investigator-initiated phase 2 study of pembrolizumab immunological response evaluation (INSPIRE). J Immunother Cancer. 2019; 7:72.56. Köhler F, Rodríguez-Paredes M. DNA methylation in epidermal differentiation, aging, and cancer. J Invest Dermatol. 2020; 140:38–47.57. Lennon AM, Buchanan AH, Kinde I, Warren A, Honushefsky A, Cohain AT, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020; 369:e. abb9601.58. GRAIL. Clinical expertise [Internet]. San Francisco (CA): GRAIL;[cited 2022 Aug 1]. Available from: https://grail.com/clinicalexpertise/.59. Liu S, Wang J. Current and future perspectives of cell-free DNA in liquid biopsy. Curr Issues Mol Biol. 2022; 44:2695–2709.60. Klein EA, Richards D, Cohn A, Tummala M, Lapham R, Cosgrove D, et al. Clinical validation of a targeted methylation-based multicancer early detection test using an independent validation set. Ann Oncol. 2021; 32:1167–1177.61. Danielson KM, Rubio R, Abderazzaq F, Das S, Wang YE. High throughput sequencing of extracellular RNA from human plasma. PLoS One. 2017; 12:e0164644.62. Vorperian SK, Moufarrej MN; Tabula Sapiens Consortium, Quake SR. Cell types of origin of the cell-free transcriptome. Nat Biotechnol. 2022; 40:855–861.63. Larson MH, Pan W, Kim HJ, Mauntz RE, Stuart SM, Pimentel M, et al. A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat Commun. 2021; 12:2357.64. Köberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, PevelingOberhag J, et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013; 49:3442–3449.65. Xu Y, Bu X, Dai C, Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour Biol. 2015; 36:4773–4776.66. Cho HJ, Kim SS, Nam JS, Kim JK, Lee JH, Kim B, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol. 2017; 41:181–189.67. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019; 88:487–514.68. Chen W, Mao Y, Liu C, Wu H, Chen S. Exosome in hepatocellular carcinoma: an update. J Cancer. 2021; 12:2526–2536.69. Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016; 74:103–141.70. Shen M, Di K, He H, Xia Y, Xie H, Huang R, et al. Progress in exosome associated tumor markers and their detection methods. Mol Biomed. 2020; 1:3.71. Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun. 2020; 11:4489.72. Wang Y, Zhang C, Zhang P, Guo G, Jiang T, Zhao X, et al. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018; 7:1670–1679.73. Ashworth TR. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Aust. 1869; 14:146–147.74. Recasens A, Munoz L. Targeting cancer cell dormancy. Trends Pharmacol Sci. 2019; 40:128–141.75. Cui K, Ou Y, Shen Y, Li S, Sun Z. Clinical value of circulating tumor cells for the diagnosis and prognosis of hepatocellular carcinoma (HCC): a systematic review and meta-analysis. Medicine (Baltimore). 2020; 99:e22242.76. Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the CellSearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010; 2010:617421.77. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014; 158:1110–1122.78. Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021; 73:422–436.79. Bankó P, Lee SY, Nagygyörgy V, Zrínyi M, Chae CH, Cho DH, et al. Technologies for circulating tumor cell separation from whole blood. J Hematol Oncol. 2019; 12:48.80. Ou H, Huang Y, Xiang L, Chen Z, Fang Y, Lin Y, et al. Circulating tumor cell phenotype indicates poor survival and recurrence after surgery for hepatocellular carcinoma. Dig Dis Sci. 2018; 63:2373–2380.81. Yu JJ, Xiao W, Dong SL, Liang HF, Zhang ZW, Zhang BX, et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer. 2018; 18:835.82. Li YM, Xu SC, Li J, Han KQ, Pi HF, Zheng L, et al. Epithelial-mesenchymal transition markers expressed in circulating tumor cells in hepatocellular carcinoma patients with different stages of disease. Cell Death Dis. 2013; 4:e831.83. Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018; 24:946–960.84. Yi B, Wu T, Zhu N, Huang Y, Yang X, Yuan L, et al. The clinical significance of CTC enrichment by GPC3-IML and its genetic analysis in hepatocellular carcinoma. J Nanobiotechnology. 2021; 19:74.85. Noh CK, Wang HJ, Kim CM, Kim J, Yoon SY, Lee GH, et al. EpCAM as a predictive marker of tumor recurrence and survival in patients who underwent surgical resection for hepatocellular carcinoma. Anticancer Res. 2018; 38:4101–4109.86. Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X, et al. Association of preoperative EpCAM circulating tumor cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer. 2016; 16:506.87. Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res. 2014; 20:4794–4805.88. Ye X, Li G, Han C, Han Q, Shang L, Su H, et al. Circulating tumor cells as a potential biomarker for postoperative clinical outcome in HBV-related hepatocellular carcinoma. Cancer Manag Res. 2018; 10:5639–5647.89. Teng PC, Agopian VG, Lin TY, You S, Zhu Y, Tseng HR, et al. Circulating tumor cells: A step toward precision medicine in hepatocellular carcinoma. J Gastroenterol Hepatol. 2022; 37:1179–1190.90. Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013; 57:1458–1468.91. Zhang J, Quadri S, Wolfgang CL, Zheng L. New development of biomarkers for gastrointestinal cancers: from neoplastic cells to tumor microenvironment. Biomedicines. 2018; 6:87.92. Abdelgawad IA. Epithelial cell adhesion molecule mRNA can be a potential marker to predict metastasis in hepatocellular carcinoma patients. Asian Pac J Cancer Prev. 2020; 21:861–866.93. Fan JL, Yang YF, Yuan CH, Chen H, Wang FB. Circulating tumor cells for predicting the prognostic of patients with hepatocellular carcinoma: a meta analysis. Cell Physiol Biochem. 2015; 37:629–640.94. Calabuig-Fariñas S, Jantus-Lewintre E, Herreros-Pomares A, Camps C. Circulating tumor cells versus circulating tumor DNA in lung cancer-which one will win? Transl Lung Cancer Res. 2016; 5:466–482.95. Exact Sciences. Frequently asked questions [Internet]. Madison (WI): Cologuard;[cited 2022 Aug 1]. Available from: https://www.cologuardhcp.com/faq.96. Li Q, He Z, Guo Y, Zhang H, George TJ, Hogan W, et al. Assessing the validity of a a priori patient-trial generalizability score using real-world data from a large clinical data research network: a colorectal cancer clinical trial case study. AMIA Annu Symp Proc. 2020; 2019:1101–1110.97. Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018; 36:1631–1641.98. Fleischhacker M, Schmidt B. Pre-analytical issues in liquid biopsy – where do we stand? J Lab Med. 2020; 44:117–142.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current Status and Prospects of Liquid Biopsy for Hepatocellular Carcinoma
- Liquid biopsy in hepatocellular carcinoma: Challenges, advances, and clinical implications
- A Concise Review of Autoantibodies to Tumor-Associated Antigens for Diagnosis of Hepatocellular Carcinoma
- Clinical Application of Circulating Tumor Cells in Gastric Cancer
- A Case of Spontaneous Regression of Hepatocellular Carcinoma after Ultrasound Guided Liver Biopsy