J Korean Med Sci.
2022 Oct;37(39):e289. 10.3346/jkms.2022.37.e289.
Evaluation of In-Hospital Cluster of COVID-19 Associated With a Patient With Prolonged Viral Shedding Using Whole-Genome Sequencing
- Affiliations
-
- 1Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Office for Infection Control, Asan Medical Center, Seoul, Korea
- 3Department of Microbiology, Institute for Viral Diseases, Vaccine Innovation Center, College of Medicine, Korea University, Seoul, Korea
- KMID: 2534027
- DOI: http://doi.org/10.3346/jkms.2022.37.e289
Abstract
- Background
Patients with hematologic malignancies may produce replication-competent virus beyond 20 days of SARS-CoV-2 infection. However, data regarding the transmission of SARS-CoV-2 from patients with prolonged viral shedding is limited.
Methods
In May 2022, four additional cases of COVID-19 were reported in a hematologic ward at a tertiary care hospital in South Korea, after an 8-week isolation of a patient with prolonged viral shedding. We performed whole-genome sequencing (WGS) of SARS-CoV-2 to evaluate the possibility of post-isolation transmission from this prolonged viral shedding.
Results
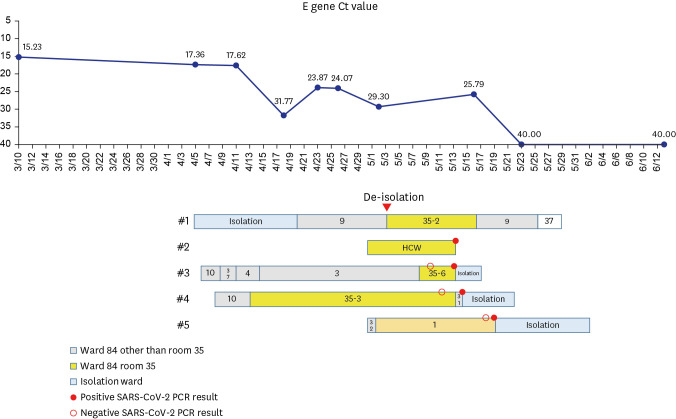
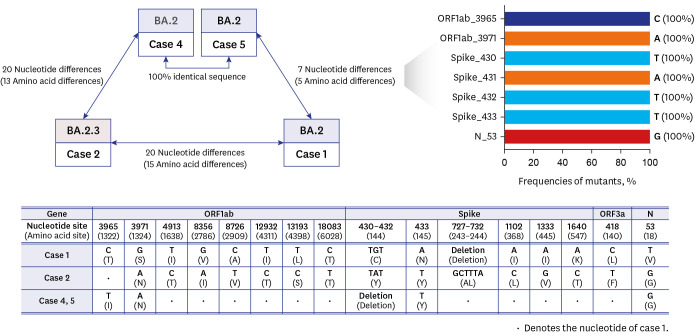
A patient (case 1) with acute myeloid leukemia was released from isolation 54 days after the diagnosis of COVID-19 based on rising Ct value of up to 29.3, and moved to a sixpatient room. On days 10 and 11 post-isolation, his doctor (case 2) and 2 patients who were his roommates (case 3, 4) had positive SARS-CoV-2 PCR results. Additionally, 16 days postisolation, another patient (case 5) in a remote room had positive SARS-CoV-2 PCR result. All the three patients were hospitalized for ≥ 14 days when they were diagnosed with SARS-CoV-2 infection. Except for case 3, the remaining 4 cases were available for WGS, which revealed that case 1 exhibited a 7 nucleotides difference in comparison to cases 4 and 5 and case 2 displayed a 20 nucleotides difference compared with case 1, while sequences of cases 4 and 5 were identical.
Conclusions
Despite the possibility of transmission from the patient with prolonged viral shedding, no evidence of the transmission of SARS-CoV-2 from the patient with prolonged positive RT-PCR using WGS was found.
Keyword
Figure
Reference
-
1. Centers for Disease Control and Prevention. Ending isolation and precautions for people with COVID-19: interim guidance. Accessed June 30, 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html .2. Aydillo T, Gonzalez-Reiche AS, Aslam S, van de Guchte A, Khan Z, Obla A, et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med. 2020; 383(26):2586–2588. PMID: 33259154.
Article3. Sung A, Bailey AL, Stewart HB, McDonald D, Wallace MA, Peacock K, et al. Isolation of SARS-CoV-2 in viral cell culture in immunocompromised patients with persistently positive RT-PCR results. Front Cell Infect Microbiol. 2022; 12:804175. PMID: 35186791.
Article4. Kim JY, Bae JY, Bae S, Cha HH, Kwon JS, Suh MH, et al. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin Microbiol Infect. 2022; 28(1):101–106. PMID: 34400343.
Article5. Jung J, Lim SY, Lee J, Bae S, Lim YJ, Hong MJ, et al. Clustering and multiple-spreading events of nosocomial severe acute respiratory syndrome coronavirus 2 infection. J Hosp Infect. 2021; 117:28–36. PMID: 34453983.
Article6. Klompas M, Baker MA, Rhee C, Tucker R, Fiumara K, Griesbach D, et al. A SARS-CoV-2 cluster in an acute care hospital. Ann Intern Med. 2021; 174(6):794–802. PMID: 33556277.
Article7. Jung J, Lee J, Park H, Lim YJ, Kim EO, Park MS, et al. Nosocomial outbreak by delta variant from a fully vaccinated patient. J Korean Med Sci. 2022; 37(17):e133. PMID: 35502502.
Article8. Czech-Sioli M, Günther T, Robitaille A, Roggenkamp H, Büttner H, Indenbirken D, et al. Integration of sequencing and epidemiological data for surveillance of SARS-CoV-2 infections in a tertiary-care hospital. Clin Infect Dis. Forthcoming. 2022; DOI: 10.1093/cid/ciac484.
Article9. Zürcher K, Abela IA, Stange M, Dupont C, Mugglin C, Egli A, et al. Alpha variant coronavirus outbreak in a nursing home despite high vaccination coverage: molecular, epidemiological and immunological studies. Clin Infect Dis. Forthcoming. 2022; DOI: 10.1093/cid/ciab1005.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Reinfection of SARS-CoV-2 Variants in Immunocompromised Patients with Prolonged or Relapsed Viral Shedding
- Viral Shedding Kinetics in Patients with COVID-19
- A Case of Whole Genome Analysis of SARSCoV-2 Using Oxford Nanopore MinION System
- Viral shedding patterns of symptomatic SARS-CoV-2 infections by periods of variant predominance and vaccination status in Gyeonggi Province, Korea
- A Case of COVID-19 in a 45-Day-Old Infant with Persistent Fecal Virus Shedding for More Than 12 Weeks