Ann Surg Treat Res.
2022 Aug;103(2):63-71. 10.4174/astr.2022.103.2.63.
Overexpression of FRAT1 protein is closely related to triple-negative breast cancer
- Affiliations
-
- 1Department of Surgery, Konkuk University School of Medicine, Seoul, Korea
- 2Research Institute of Medical Science, Konkuk University School of Medicine, Seoul, Korea
- 3Pathology Center, Seegene Medical Foundation, Seoul, Korea
- 4Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
- KMID: 2532360
- DOI: http://doi.org/10.4174/astr.2022.103.2.63
Abstract
- Purpose
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with a poor prognosis and a lack of targeted therapy. Overexpression of FRAT1 is thought to be associated with this aggressive subtype of cancer. Here, we performed a comprehensive analysis and assessed the association between overexpression of FRAT1 and TNBC.
Methods
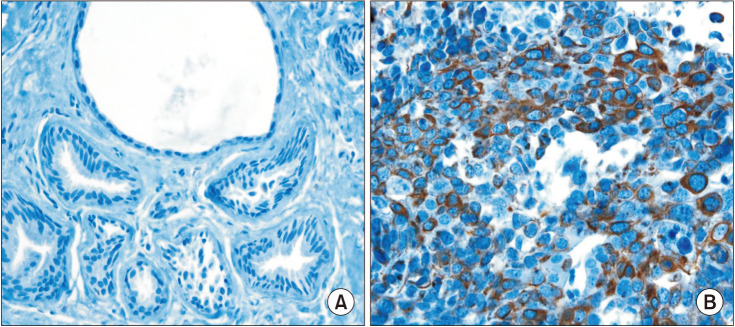
First, using different web-based bioinformatics platforms (TIMER 2.0, UALCAN, and GEPIA 2), the expression of FRAT1 was assessed. Then, the expression of the FRAT1 protein and hormone receptors and HER2 status were assessed by immunohistochemical analysis. For samples of tumors with equivocal immunoreactivity, we performed silver in situ hybridization of the HER2 gene to determine an accurate HER2 status. Next, we used the R package and bc-GenExMiner 4.8 to analyze the relationship between FRAT1 expression and clinicopathological parameters in breast cancer patients. Finally, we determined the relationship between FRAT1 overexpression and prognosis in patients.
Results
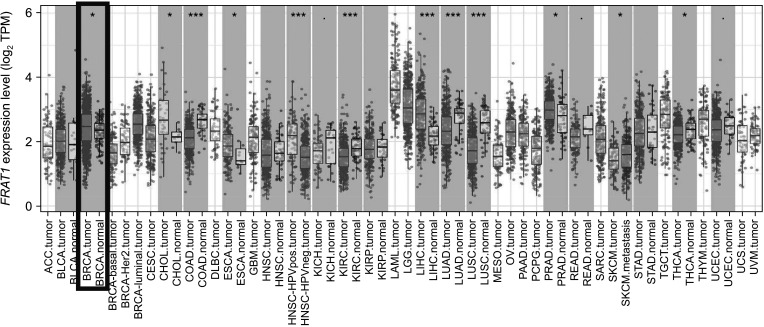
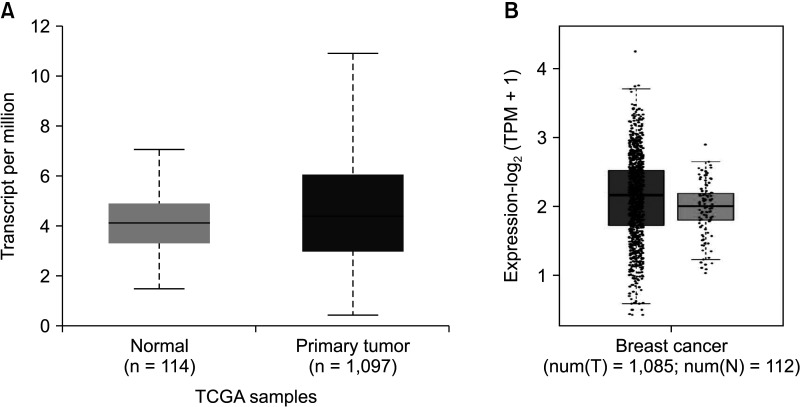
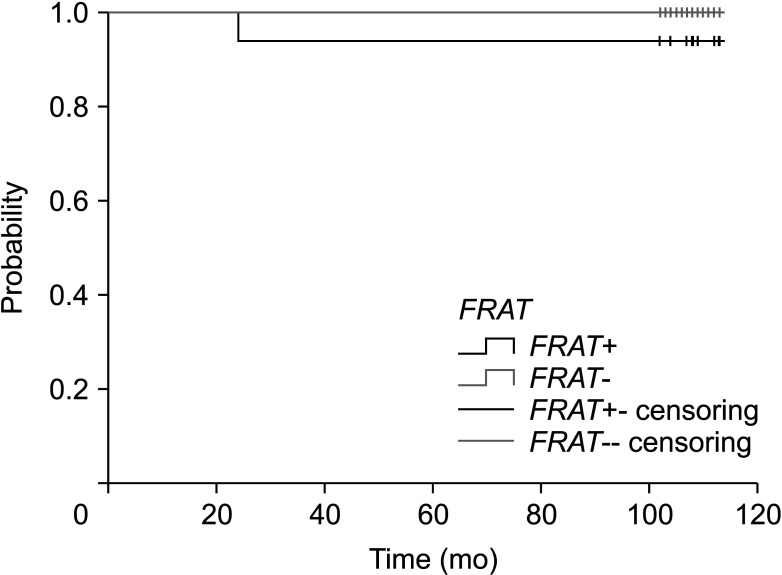
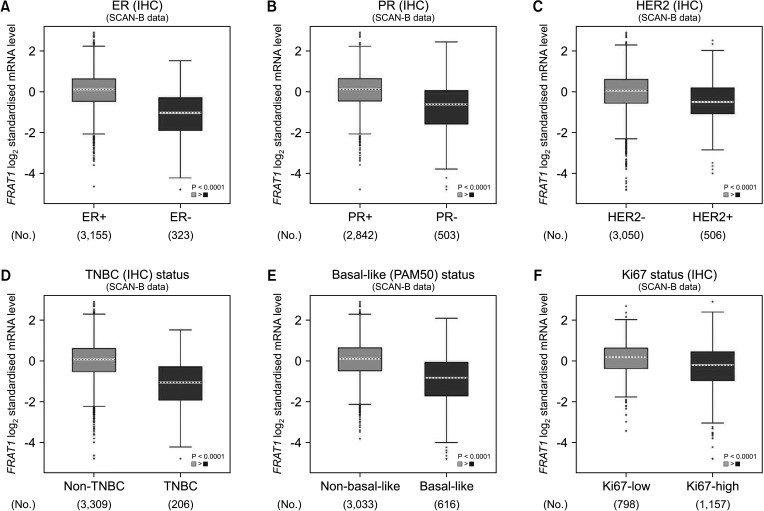
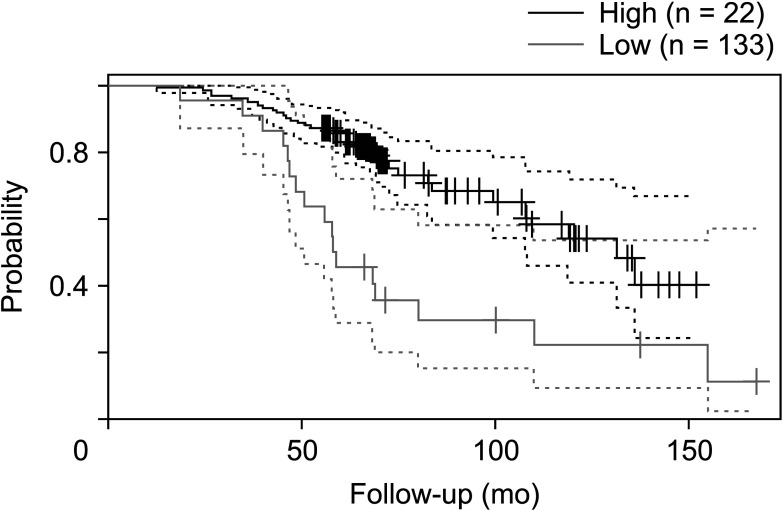
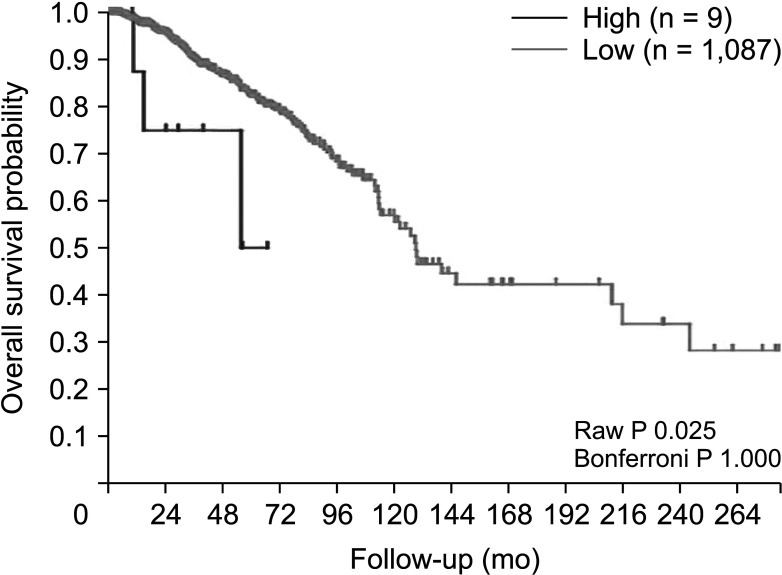
The expression of FRAT1 in breast cancer tissues is significantly higher than in normal tissue. FRAT1 expression was significantly related to worse overall survival (P < 0.05) and was correlated with these clinicopathological features: T stage, N stage, age, high histologic grade, estrogen receptor status, progesterone receptor status, Her-2 status, TNBC status, basal-like status, CK5/6 status, and Ki67 status.
Conclusion
FRAT1 was overexpressed in breast cancer compared to normal tissue, and it may be involved in the progression of breast cancer malignancy. This study provides suggestive evidence of the prognostic role of FRAT1 in breast cancer and the therapeutic target for TNBC.
Keyword
Figure
Reference
-
1. Althuis MD, Dozier JM, Anderson WF, Devesa SS, Brinton LA. Global trends in breast cancer incidence and mortality 1973-1997. Int J Epidemiol. 2005; 34:405–412. PMID: 15737977.
Article2. Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol. 2010; 23 Suppl 2:S60–S64. PMID: 20436504.
Article3. Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011; 121:3786–3788. PMID: 21965334.
Article4. Liao C, Zhang Y, Fan C, Herring LE, Liu J, Locasale JW, et al. Identification of BBOX1 as a therapeutic target in triple-negative breast cancer. Cancer Discov. 2020; 10:1706–1721. PMID: 32690540.
Article5. Anders CK, Carey LA. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin Breast Cancer. 2009; 9(Suppl 2):S73–S81. PMID: 19596646.
Article6. Metzger-Filho O, Tutt A, de Azambuja E, Saini KS, Viale G, Loi S, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol. 2012; 30:1879–1887. PMID: 22454417.
Article7. Saitoh T, Katoh M. FRAT1 and FRAT2, clustered in human chromosome 10q24.1 region, are up-regulated in gastric cancer. Int J Oncol. 2001; 19:311–315. PMID: 11445844.
Article8. Zhang Y, Yu JH, Lin XY, Miao Y, Han Y, Fan CF, et al. Overexpression of Frat1 correlates with malignant phenotype and advanced stage in human non-small cell lung cancer. Virchows Arch. 2011; 459:255–263. PMID: 21818639.
Article9. Hagen T, Cross DA, Culbert AA, West A, Frame S, Morrice N, et al. FRAT1, a substrate-specific regulator of glycogen synthase kinase-3 activity, is a cellular substrate of protein kinase A. J Biol Chem. 2006; 281:35021–35029. PMID: 16982607.
Article10. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: update on effectors and inhibitors. Cancer Treat Rev. 2018; 62:50–60. PMID: 29169144.
Article11. Guo G, Mao X, Wang P, Liu B, Zhang X, Jiang X, et al. The expression profile of FRAT1 in human gliomas. Brain Res. 2010; 1320:152–158. PMID: 20096670.
Article12. Wang Y, Hewitt SM, Liu S, Zhou X, Zhu H, Zhou C, et al. Tissue microarray analysis of human FRAT1 expression and its correlation with the subcellular localisation of beta-catenin in ovarian tumours. Br J Cancer. 2006; 94:686–691. PMID: 16479254.
Article13. Wang Y, Liu S, Zhu H, Zhang W, Zhang G, Zhou X, et al. FRAT1 overexpression leads to aberrant activation of beta-catenin/TCF pathway in esophageal squamous cell carcinoma. Int J Cancer. 2008; 123:561–568. PMID: 18498136.
Article14. Zhu K, Guo J, Wang H, Yu W. FRAT1 expression regulates proliferation in colon cancer cells. Oncol Lett. 2016; 12:4761–4766. PMID: 28101222.
Article15. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19(1A):A68–A77. PMID: 25691825.16. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020; 48(W1):W509–W514. PMID: 32442275.
Article17. Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017; 19:649–658. PMID: 28732212.
Article18. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017; 45(W1):W98–W102. PMID: 28407145.
Article19. Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012; 131:765–775. PMID: 21452023.
Article20. Hashmi AA, Naz S, Hashmi SK, Hussain ZF, Irfan M, Bakar SM, et al. Cytokeratin 5/6 and cytokeratin 8/18 expression in triple negative breast cancers: clinicopathologic significance in South-Asian population. BMC Res Notes. 2018; 11:372. PMID: 29884220.
Article21. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016; 13:674–690. PMID: 27184417.
Article22. Saitoh T, Mine T, Katoh M. Molecular cloning and expression of proto-oncogene FRAT1 in human cancer. Int J Oncol. 2002; 20:785–789. PMID: 11894125.
Article23. Zhang Y, Han Y, Zheng R, Yu JH, Miao Y, Wang L, et al. Expression of Frat1 correlates with expression of β-catenin and is associated with a poor clinical outcome in human SCC and AC. Tumour Biol. 2012; 33:1437–1444. PMID: 22528942.
Article24. Yost C, Farr GH 3rd, Pierce SB, Ferkey DM, Chen MM, Kimelman D. GBP, an inhibitor of GSK-3, is implicated in Xenopus development and oncogenesis. Cell. 1998; 93:1031–1041. PMID: 9635432.
Article25. Thomas GM, Frame S, Goedert M, Nathke I, Polakis P, Cohen P. A GSK3-binding peptide from FRAT1 selectively inhibits the GSK3-catalysed phosphorylation of axin and beta-catenin. FEBS Lett. 1999; 458:247–251. PMID: 10481074.
Article26. Li L, Yuan H, Weaver CD, Mao J, Farr GH 3rd, Sussman DJ, et al. Axin and Frat1 interact with dvl and GSK, bridging Dvl to GSK in Wnt-mediated regulation of LEF-1. EMBO J. 1999; 18:4233–4240. PMID: 10428961.
Article27. Walf-Vorderwülbecke V, de Boer J, Horton SJ, van Amerongen R, Proost N, Berns A, et al. Frat2 mediates the oncogenic activation of Rac by MLL fusions. Blood. 2012; 120:4819–4828. PMID: 23074275.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- LncRNA DLG1-AS1 Promotes Cancer Cell Proliferation in Triple Negative Breast Cancer by Downregulating miR-203
- The expression of Rab5 and its effect on invasion, migration and exosome secretion in triple negative breast cancer
- Clinicopathologic Characteristics and Prognosis of Early Stage Triple Negative Breast Cancer: Comparison with Non-triple Negative Group
- Treatment Outcomes of Weakly Positive Hormone Receptor Breast Cancer and Triple-Negative Breast Cancer
- Fear of Cancer Recurrence and Unmet Needs in Triple Negative Breast Cancer Survivors