Kosin Med J.
2022 Jun;37(2):154-162. 10.7180/kmj.22.114.
Performance comparison between Elecsys Anti-SARS-CoV-2 and Anti-SARS-CoV-2 S and Atellica IM SARS-CoV-2 Total and SARS-CoV-2 IgG assays
- Affiliations
-
- 1Department of Laboratory Medicine, Hallym University Kangnam Sacred Heart Hospital, Hallym University College of Medicine, Seoul, Korea
- 2Department of Pediatrics, Kosin University Gospel Hospital, Kosin University College of Medicine, Busan, Korea
- 3Department of Laboratory Medicine, Kosin University Gospel Hospital, Kosin University College of Medicine, Busan, Korea
- KMID: 2532046
- DOI: http://doi.org/10.7180/kmj.22.114
Abstract
- Background
Although serological severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) tests from several manufacturers have been introduced in South Korea and some are commercially available, the performance of these test kits has not yet been sufficiently validated. Therefore, we compared the performance of Elecsys Anti-SARS-CoV-2 (ACOV2) and Anti-SARS-CoV-2 S (ACOV2S) and Atellica IM SARS-CoV-2 Total (COV2T) and SARS-CoV-2 IgG (sCOVG) serological tests in this study.
Methods
A total of 186 patient samples were used. For each test, we analyzed the positive rate of serological antibody tests, precision, linearity, and agreement among the four assays.
Results
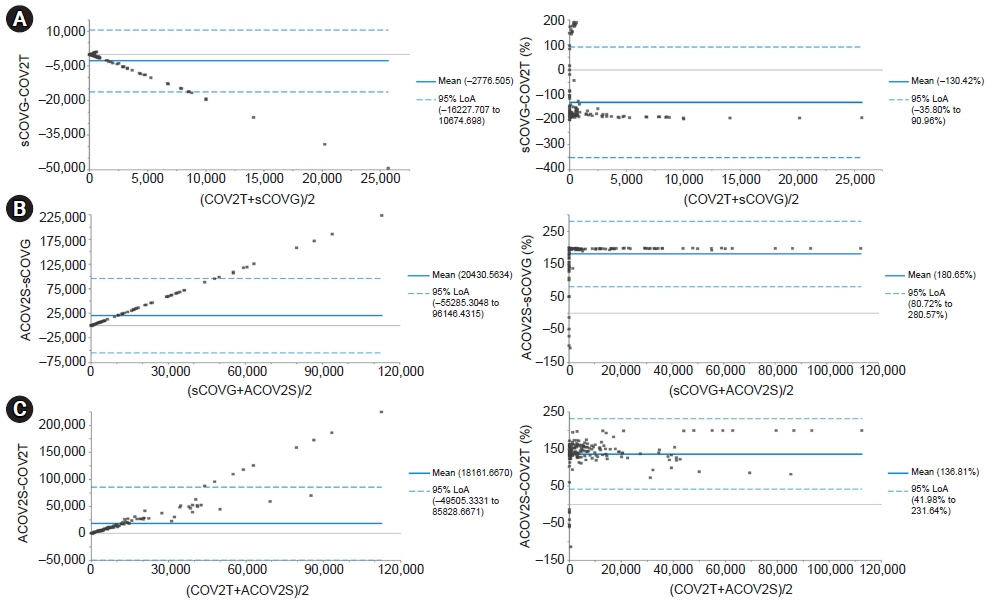
The positive rates of COV2T, sCOVG, and ACOV2S were high (81.7%–89.2%) in total, with those for ACOV2S being the highest, while those of ACOV2 were as low as 44.6%. This may be related to the high completion rate of vaccination in Korea. The repeatability and within-laboratory coefficients of variation were within the claimed allowable imprecision; however, further research is needed to establish an allowable imprecision at low concentrations. COV2T showed a linear fit, whereas sCOVG and ACOV2S were appropriately modeled with a nonlinear fit. Good agreement was found among COV2T, sCOVG, and ACOV2S; however, the agreement between ACOV2 and any one of the other methods was poor.
Conclusions
Considering the different antigens used in serological SARS-CoV-2 antibody assays, the performance of the tested assays is thought to show no significant difference for the qualitative detection of antibodies to SARS-CoV-2.
Keyword
Figure
Cited by 2 articles
-
Evaluation of automated calibration and quality control processes using the Aptio total laboratory automation system
Namhee Kim, Yein Kim, Jeongeun Park, Jungsoo Choi, Hyunyong Hwang
Kosin Med J. 2022;37(4):342-353. doi: 10.7180/kmj.22.144.Comparative analysis of Access PCT and Elecsys BRAHMS PCT assays for procalcitonin measurements
Hyunji Choi, Sang-Shin Lee, Hyunyong Hwang
Kosin Med J. 2024;39(4):272-280. doi: 10.7180/kmj.24.155.
Reference
-
References
1. Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020; 16:1686–97.
Article2. Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020; 12:e7423.
Article3. Safiabadi Tali SH, LeBlanc JJ, Sadiq Z, Oyewunmi OD, Camargo C, Nikpour B, et al. Tools and techniques for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)/COVID-19 detection. Clin Microbiol Rev. 2021; 34:e00228–20.
Article4. Jolobe OMP. What are the criteria for asymptomatic status? QJM. 2021; 114:351–2.
Article5. LeBlanc JJ, Gubbay JB, Li Y, Needle R, Arneson SR, Marcino D, et al. Real-time PCR-based SARS-CoV-2 detection in Canadian laboratories. J Clin Virol. 2020; 128:104433.
Article6. Bhimraj A, Morgan RL, Shumaker AH, Lavergne V, Baden L, Cheng VC, et al. Infectious Diseases Society of America Guidelines on the treatment and management of patients with COVID-19. Clin Infect Dis. 2020 Apr 27 [Epub]. https://doi.org/10.1093/cid/ciaa478.
Article7. Stegeman I, Ochodo EA, Guleid F, Holtman GA, Yang B, Davenport C, et al. Routine laboratory testing to determine if a patient has COVID-19. Cochrane Database Syst Rev. 2020; 11:CD013787.
Article8. Nakagama Y, Komase Y, Candray K, Nakagama S, Sano F, Tsuchida T, et al. Serological testing reveals the hidden COVID-19 burden among health care workers experiencing a SARS-CoV-2 nosocomial outbreak. Microbiol Spectr. 2021; 9:e0108221.
Article9. Moura DTH, McCarty TR, Ribeiro IB, Funari MP, Oliveira PVAG, Miranda Neto AA, et al. Diagnostic characteristics of serological-based COVID-19 testing: a systematic review and meta-analysis. Clinics (Sao Paulo). 2020; 75:e2212.
Article10. Kadam SB, Sukhramani GS, Bishnoi P, Pable AA, Barvkar VT. SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights. J Basic Microbiol. 2021; 61:180–202.
Article11. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020; 5:562–9.
Article12. Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020; 12:8.
Article13. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–3.
Article14. Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S. The spike protein of SARS-CoV: a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009; 7:226–36.
Article15. Saputri DS, Li S, van Eerden FJ, Rozewicki J, Xu Z, Ismanto HS, et al. Flexible, functional, and familiar: characteristics of SARS-CoV-2 spike protein evolution. Front Microbiol. 2020; 11:2112.
Article16. Hwang YC, Lu RM, Su SC, Chiang PY, Ko SH, Ke FY, et al. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J Biomed Sci. 2022; 29:1.
Article17. Giavarina D, Carta M. Improvements and limits of anti SARS-CoV-2 antibodies assays by WHO (NIBSC 20/136) standardization. Diagnosis (Berl). 2021; 9:274–9.
Article18. Aguirre JJ, Ness K; Algeciras-Schimnich A. Application of the CLSI EP15-A3 guideline as an alternative troubleshooting tool for verification of assay precision. Am J Clin Pathol. 2019; 152(Suppl 1):S88.
Article19. Tholen DW, Kroll M, Astles JR, Caffo AL, Happe TM, Krouwer J, et al. Evaluation of the linearity of quantitative measurement procedures: a statistical approach; approved guideline. Wayne: Clinical and Laboratory Standards Institute;2003.20. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977; 33:159–74.
Article21. Nittari G, Pallotta G, Amenta F, Tayebati SK. Current pharmacological treatments for SARS-COV-2: a narrative review. Eur J Pharmacol. 2020; 882:173328.
Article22. Trougakos IP, Stamatelopoulos K, Terpos E, Tsitsilonis OE, Aivalioti E, Paraskevis D, et al. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J Biomed Sci. 2021; 28:9.
Article23. Tiwari N, Upadhyay J, Ansari MN, Joshi R. Novel β-coronavirus (SARS-CoV-2): current and future aspects of pharmacological treatments. Saudi Pharm J. 2020; 28:1243–52.
Article24. Kim MK, Lee B, Choi YY, Um J, Lee KS, Sung HK, et al. Clinical characteristics of 40 patients infected with the SARS-CoV-2 Omicron variant in Korea. J Korean Med Sci. 2022; 37:e31.
Article25. Leuzinger K, Osthoff M, Drager S, Pargger H, Siegemund M, Bassetti S, et al. Comparing immunoassays for SARS-CoV-2 antibody detection in patients with and without laboratory-confirmed SARS-CoV-2 infection. J Clin Microbiol. 2021; 59:e0138121.
Article26. Kim MA, Lee YW, Kim SR, Kim JH, Min TK, Park HS, et al. COVID-19 vaccine-associated anaphylaxis and allergic reactions: consensus statements of the KAAACI Urticaria/Angioedema/Anaphylaxis Working Group. Allergy Asthma Immunol Res. 2021; 13:526–44.
Article27. Igawa G, Ai T, Yamamoto T, Ito K, Nojiri S, Saito K, et al. Antibody response and seroprevalence in healthcare workers after the BNT162b2 vaccination in a University Hospital at Tokyo. Sci Rep. 2022; 12:8707.
Article28. Florin L, Maelegheer K, Vandewal W, Bernard D, Robbrecht J. Performance evaluation of the Siemens SARS-CoV-2 Total Antibody and IgG Antibody Test. Lab Med. 2021; 52:e147–3.
Article29. Haselmann V, Kittel M, Gerhards C, Thiaucourt M, Eichner R, Costina V, et al. Comparison of test performance of commercial anti-SARS-CoV-2 immunoassays in serum and plasma samples. Clin Chim Acta. 2020; 510:73–8.
Article30. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986; 1:307–10.
Article31. Giavarina D. Understanding Bland Altman analysis. Biochem Med (Zagreb). 2015; 25:141–51.
Article32. Gao J, Quan L. Current status of diagnostic testing for SARS-CoV-2 infection and future developments: a review. Med Sci Monit. 2020; 26:e928552.
Article33. Xu X, Sun J, Nie S, Li H, Kong Y, Liang M, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat Med. 2020; 26:1193–5.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Understandings and Prospects of Laboratory Diagnosis of SARS-CoV-2
- Challenges of Scaling Up SARS-CoV-2 Rapid Antigen Tests
- Changes in SARS-CoV-2 antibody titers 6 months after the booster dose of BNT162b2 COVID-19 vaccine among health care workers
- Anti-SARS-CoV-2 spike antibody response to the third dose of BNT162b2 mRNA COVID-19 vaccine and associated factors in Japanese hemodialysis patients
- SARS-CoV-2-Specific T Cell Responses in Patients with COVID-19 and Unexposed Individuals