Brain Tumor Res Treat.
2022 Apr;10(2):76-82. 10.14791/btrt.2021.0032.
Digital Pathology and Artificial Intelligence Applications in Pathology
- Affiliations
-
- 1Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- KMID: 2529438
- DOI: http://doi.org/10.14791/btrt.2021.0032
Abstract
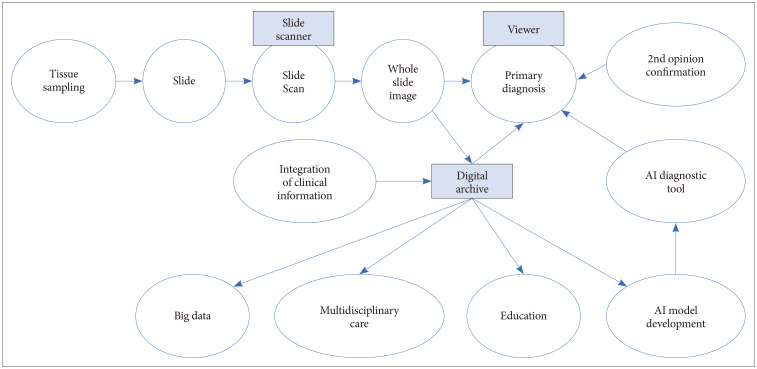
- Digital pathology is revolutionizing pathology. The introduction of digital pathology made it possible to comprehensively change the pathology diagnosis workflow, apply and develop pathological artificial intelligence (AI) models, generate pathological big data, and perform telepathology. AI algorithms, including machine learning and deep learning, are used for the detection, segmentation, registration, processing, and classification of digitized pathological images. Pathological AI algorithms can be helpfully utilized for diagnostic screening, morphometric analysis of biomarkers, the discovery of new meanings of prognosis and therapeutic response in pathological images, and improvement of diagnostic efficiency. In order to develop a successful pathological AI model, it is necessary to consider the selection of a suitable type of image for a subject, utilization of big data repositories, the setting of an effective annotation strategy, image standardization, and color normalization. This review will elaborate on the advantages and perspectives of digital pathology, AI-based approaches, the applications in pathology, and considerations and challenges in the development of pathological AI models.
Figure
Reference
-
1. Nam S, Chong Y, Jung CK, Kwak TY, Lee JY, Park J, et al. Introduction to digital pathology and computer-aided pathology. J Pathol Transl Med. 2020; 54:125–134. PMID: 32045965.2. Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019; 16:703–715. PMID: 31399699.3. Jahn SW, Plass M, Moinfar F. Digital pathology: advantages, limitations and emerging perspectives. J Clin Med. 2020; 9:3697.4. Barisoni L, Lafata KJ, Hewitt SM, Madabhushi A, Balis UGJ. Digital pathology and computational image analysis in nephropathology. Nat Rev Nephrol. 2020; 16:669–685. PMID: 32848206.5. Mukhopadhyay S, Feldman MD, Abels E, Ashfaq R, Beltaifa S, Cacciabeve NG, et al. Whole slide imaging versus microscopy for primary diagnosis in surgical pathology: a multicenter blinded randomized noninferiority study of 1992 cases (pivotal study). Am J Surg Pathol. 2018; 42:39–52. PMID: 28961557.6. Goacher E, Randell R, Williams B, Treanor D. The diagnostic concordance of whole slide imaging and light microscopy: a systematic review. Arch Pathol Lab Med. 2017; 141:151–161. PMID: 27399211.7. Snead DR, Tsang YW, Meskiri A, Kimani PK, Crossman R, Rajpoot NM, et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology. 2016; 68:1063–1072. PMID: 26409165.8. Azam AS, Miligy IM, Kimani PK, Maqbool H, Hewitt K, Rajpoot NM, et al. Diagnostic concordance and discordance in digital pathology: a systematic review and meta-analysis. J Clin Pathol. 2021; 74:448–455. PMID: 32934103.9. van Diest PJ, Huisman A, van Ekris J, Meijer J, Willems S, Hofhuis H, et al. Pathology image exchange: the Dutch digital pathology platform for exchange of whole-slide images for efficient teleconsultation, telerevision, and virtual expert panels. JCO Clin Cancer Inform. 2019; 3:1–7.10. Asa SL, Bodén AC, Treanor D, Jarkman S, Lundström C, Pantanowitz L. 2020 vision of digital pathology in action. J Pathol Inform. 2019; 10:27. PMID: 31516758.11. Boyce BF. Whole slide imaging: uses and limitations for surgical pathology and teaching. Biotech Histochem. 2015; 90:321–330. PMID: 25901738.12. Alami H, Fortin JP, Gagnon MP, Pollender H, Têtu B, Tanguay F. The challenges of a complex and innovative telehealth project: a qualitative evaluation of the Eastern Quebec Telepathology Network. Int J Health Policy Manag. 2018; 7:421–432. PMID: 29764106.13. Evans AJ, Salama ME, Henricks WH, Pantanowitz L. Implementation of whole slide imaging for clinical purposes: issues to consider from the perspective of early adopters. Arch Pathol Lab Med. 2017; 141:944–959. PMID: 28440660.14. Turnquist C, Roberts-Gant S, Hemsworth H, White K, Browning L, Rees G, et al. On the edge of a digital pathology transformation: views from a cellular pathology laboratory focus group. J Pathol Inform. 2019; 10:37. PMID: 31897354.15. Abels E, Pantanowitz L, Aeffner F, Zarella MD, van der Laak J, Bui MM, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019; 249:286–294. PMID: 31355445.16. Klette R. Concise computer vision. London: Springer;2014.17. Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press;2016.18. Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: a comprehensive tutorial with selected use cases. J Pathol Inform. 2016; 7:29. PMID: 27563488.19. Holten-Rossing H, Talman MM, Jylling AMB, Laenkholm AV, Kristensson M, Vainer B. Application of automated image analysis reduces the workload of manual screening of sentinel lymph node biopsies in breast cancer. Histopathology. 2017; 71:866–873. PMID: 28677240.20. Stephens K. FDA authorizes prostate AI software [Internet]. Overland Park: AXIS Imaging News;2021. 09. 22. Accessed Dec 12, 2021. at https://axisimagingnews.com/radiology-products/radiology-software/ai-machine-learning/fda-authorizes-prostate-ai-software.21. Echle A, Grabsch HI, Quirke P, van den Brandt PA, West NP, Hutchins GGA, et al. Clinical-grade detection of microsatellite instability in colorectal tumors by deep learning. Gastroenterology. 2020; 159:1406–1416.e11. PMID: 32562722.22. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018; 24:1559–1567. PMID: 30224757.23. Lu MY, Zhao M, Shady M, Lipkova J, Chen TY, Williamson DFK, et al. Deep learning-based computational pathology predicts origins for cancers of unknown primary. arXiv [Preprint]. 2020; cited 2021 Dec 20. Available at: . DOI: 10.48550/arXiv.2006.13932.24. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021; 23:1231–1251. PMID: 34185076.25. Jain KK. A critical overview of targeted therapies for glioblastoma. Front Oncol. 2018; 8:419. PMID: 30374421.26. Zhou C, Jin Y, Chen Y, Huang S, Huang R, Wang Y, et al. Histopathology classification and localization of colorectal cancer using global labels by weakly supervised deep learning. Comput Med Imaging Graph. 2021; 88:101861. PMID: 33497891.27. Jin L, Shi F, Chun Q, Chen H, Ma Y, Wu S, et al. Artificial intelligence neuropathologist for glioma classification using deep learning on hematoxylin and eosin stained slide images and molecular markers. Neuro Oncol. 2021; 23:44–52. PMID: 32663285.28. McAvoy M, Prieto PC, Kaczmarzyk JR, Fernández IS, McNulty J, Smith T, et al. Classification of glioblastoma versus primary central nervous system lymphoma using convolutional neural networks. Sci Rep. 2021; 11:15219. PMID: 34312463.29. Harder N, Schönmeyer R, Nekolla K, Meier A, Brieu N, Vanegas C, et al. Automatic discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Sci Rep. 2019; 9:7449. PMID: 31092853.30. Brockmoeller S, Echle A, Ghaffari Laleh N, Eiholm S, Malmstrøm ML, Plato Kuhlmann T, et al. Deep learning identifies inflamed fat as a risk factor for lymph node metastasis in early colorectal cancer. J Pathol. 2022; 256:269–281. PMID: 34738636.31. Yao J, Wang S, Zhu X, Huang J. Imaging biomarker discovery for lung cancer survival prediction. In : Proceedings of the 19th International Conference on Medical Image Computing and Computer-Assisted Intervention; 2016 Oct 17-21; Athens, Greece. Springer;2016. p. 649–657.32. Jiang S, Zanazzi GJ, Hassanpour S. Predicting prognosis and IDH mutation status for patients with lower-grade gliomas using whole slide images. Sci Rep. 2021; 11:16849. PMID: 34413349.33. Rathore S, Niazi T, Iftikhar MA, Chaddad A. Glioma grading via analysis of digital pathology images using machine learning. Cancers (Basel). 2020; 12:578.34. Zadeh Shirazi A, McDonnell MD, Fornaciari E, Bagherian NS, Scheer KG, Samuel MS, et al. A deep convolutional neural network for segmentation of whole-slide pathology images identifies novel tumour cell-perivascular niche interactions that are associated with poor survival in glioblastoma. Br J Cancer. 2021; 125:337–350. PMID: 33927352.35. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015; 19:68–77.36. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018; 173:400–416.e11. PMID: 29625055.37. Thorstenson S, Molin J, Lundström C. Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: digital pathology experiences 2006-2013. J Pathol Inform. 2014; 5:14. PMID: 24843825.38. Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019; 25:1301–1309. PMID: 31308507.39. Settles B. Active learning literature survey. Computer sciences technical report. Madison: University of Wisconsin - Madison;2009. Report no. 1648.40. Wen S, Kurc TM, Hou L, Saltz JH, Gupta RR, Batiste R, et al. Comparison of different classifiers with active learning to support quality control in nucleus segmentation in pathology images. AMIA Jt Summits Transl Sci Proc. 2018; 2018:227–236.41. Herrmann MD, Clunie DA, Fedorov A, Doyle SW, Pieper S, Klepeis V, et al. Implementing the DICOM standard for digital pathology. J Pathol Inform. 2018; 9:37. PMID: 30533276.42. Vahadane A, Peng T, Sethi A, Albarqouni S, Wang L, Baust M, et al. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans Med Imaging. 2016; 35:1962–1971. PMID: 27164577.43. Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001; 23:291–299. PMID: 11531144.44. Reinhard E, Adhikhmin M, Gooch B, Shirley P. Color transfer between images. IEEE Comput Graph Appl. 2001; 21:34–41.45. Zhou N, Cai D, Han X, Yao J. Enhanced cycle-consistent generative adversarial network for color normalization of H&E stained images. In : Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019; 2019 Oct 13-17; Shenzhen, China. Springer;2019. p. 694–702.