Cancer Res Treat.
2022 Apr;54(2):383-395. 10.4143/crt.2021.759.
Radiation Response Prediction Model Based on Integrated Clinical and Genomic Data Analysis
- Affiliations
-
- 1Department of Radiation Oncology, Seoul National University Bundang Hospital, Seongnam, Korea
- 1Department of Radiation Oncology, Seoul National University Bundang Hospital, Seongnam, Korea
- 2Department of Radiation Oncology, Seoul National University College of Medicine, Seoul, Korea
- 2Department of Radiation Oncology, Seoul National University College of Medicine, Seoul, Korea
- 3Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea
- 3Graduate School of Medical Science and Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea
- 4Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- 4Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- 5Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea
- 5Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea
- 6Department of Biomedical Sciences, Seoul National University College of Medicine, Seoul, Korea
- 6Department of Biomedical Sciences, Seoul National University College of Medicine, Seoul, Korea
- 7Institute of Radiation Medicine, Medical Research Center, Seoul National University, Seoul, Korea
- 7Institute of Radiation Medicine, Medical Research Center, Seoul National University, Seoul, Korea
- KMID: 2528208
- DOI: http://doi.org/10.4143/crt.2021.759
Abstract
- Purpose
The value of the genomic profiling by targeted gene-sequencing on radiation therapy response prediction was evaluated through integrated analysis including clinical information. Radiation response prediction model was constructed based on the analyzed findings.
Materials and Methods
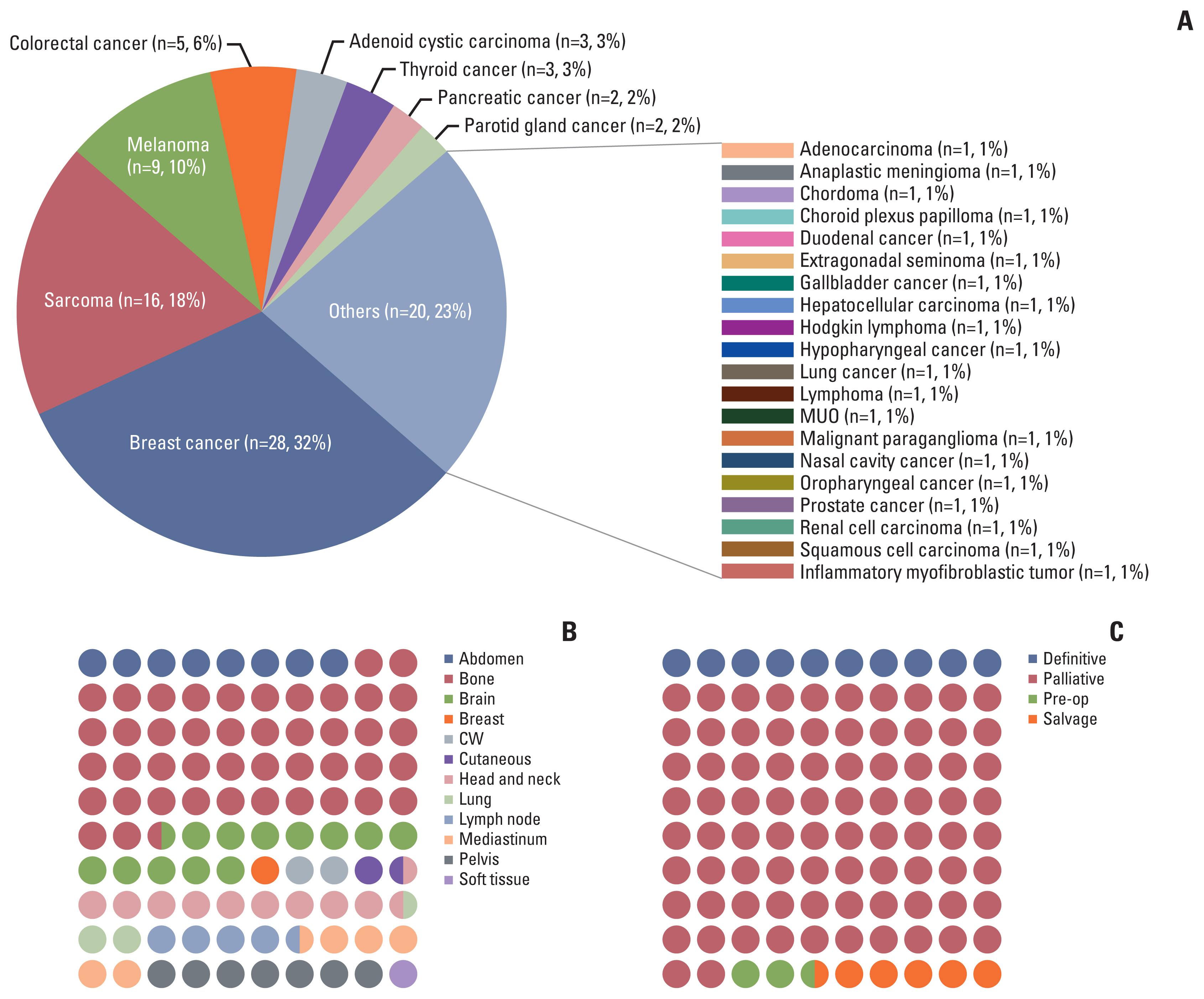
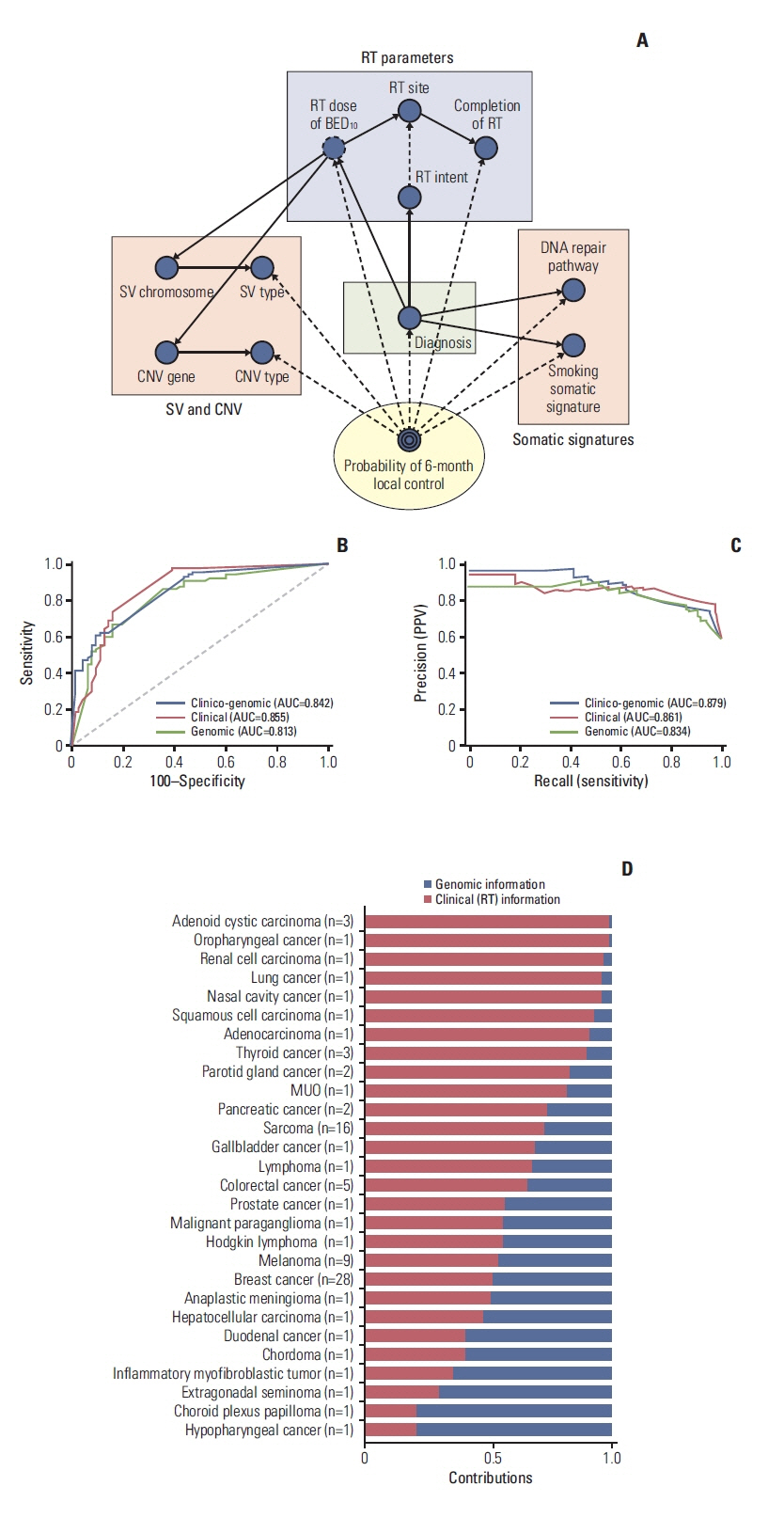
Patients who had the tumor sequenced using institutional cancer panel after informed consent and received radiotherapy for the measurable disease served as the target cohort. Patients with irradiated tumor locally controlled for more than 6 months after radiotherapy were defined as the durable local control (DLC) group, otherwise, non-durable local control (NDLC) group. Significant genomic factors and domain knowledge were used to develop the Bayesian Network model to predict radiotherapy response.
Results
Altogether, 88 patients were collected for analysis. Of those, 41 (43.6%) and 47 (54.4%) patients were classified as the NDLC and DLC group, respectively. Somatic mutations of NOTCH2 and BCL were enriched in the NDLC group, whereas, mutations of CHEK2, MSH2, and NOTCH1 were more frequently found in the DLC group. Altered DNA repair pathway was associated with better local failure–free survival (hazard ratio, 0.40; 95% confidence interval, 0.19 to 0.86; p=0.014). Smoking somatic signature was found more frequently in the DLC group. Area under the receiver operating characteristic curve of the Bayesian network model predicting probability of 6-month local control was 0.83.
Conclusion
Durable radiation response was associated with alterations of DNA repair pathway and smoking somatic signature. Bayesian network model could provide helpful insights for high precision radiotherapy. However, these findings should be verified in prospective cohort for further individualization.
Keyword
Figure
Reference
-
References
1. Hyman DM, Taylor BS, Baselga J. Implementing genome-driven oncology. Cell. 2017; 168:584–99.
Article2. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017; 23:703–13.3. Mateo J, Chakravarty D, Dienstmann R, Jezdic S, Gonzalez-Perez A, Lopez-Bigas N, et al. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Ann Oncol. 2018; 29:1895–902.
Article4. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017; 19:4–23.
Article5. Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Phys. 2009; 74:1323–31.
Article6. Bergom C, West CM, Higginson DS, Abazeed ME, Arun B, Bentzen SM, et al. The implications of genetic testing on radiation therapy decisions: a guide for radiation oncologists. Int J Radiat Oncol Biol Phys. 2019; 105:698–712.
Article7. Forker LJ, Choudhury A, Kiltie AE. Biomarkers of tumour radiosensitivity and predicting benefit from radiotherapy. Clin Oncol (R Coll Radiol). 2015; 27:561–9.
Article8. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009; 45:228–47.
Article9. Suh J, Jeong CW, Choi S, Ku JH, Kim HH, Kim K, et al. Sharing the initial experience of pan-cancer panel analysis in high-risk renal cell carcinoma in the Korean population. BMC Urol. 2020; 20:125.
Article10. Park C, Kim M, Kim MJ, Kim H, Ock CY, Keam B, et al. Clinical application of next-generation sequencing-based panel to BRAF wild-type advanced melanoma identifies key oncogenic alterations and therapeutic strategies. Mol Cancer Ther. 2020; 19:937–44.11. Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012; 28:1838–44.
Article12. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43:11.0.1–11.10.33.13. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016; 17:31.
Article14. Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018; 28:1747–56.
Article15. Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014; 30:2811–2.
Article16. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988; 44:837–45.
Article17. Laufer I, Rubin DG, Lis E, Cox BW, Stubblefield MD, Yamada Y, et al. The NOMS framework: approach to the treatment of spinal metastatic tumors. Oncologist. 2013; 18:744–51.
Article18. Husmeier D, Dybowski R, Roberts S. Probabilistic modeling in bioinformatics and medical informatics. London: Springer;London: 2005.19. Luo Y, McShan D, Ray D, Matuszak M, Jolly S, Lawrence T, et al. Development of a fully cross-validated Bayesian network approach for local control prediction in lung cancer. IEEE Trans Radiat Plasma Med Sci. 2019; 3:232–41.
Article20. Luo Y, El Naqa I, McShan DL, Ray D, Lohse I, Matuszak MM, et al. Unraveling biophysical interactions of radiation pneumonitis in non-small-cell lung cancer via Bayesian network analysis. Radiother Oncol. 2017; 123:85–92.
Article21. Ozenne B, Subtil F, Maucort-Boulch D. The precision-recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J Clin Epidemiol. 2015; 68:855–9.
Article22. Oh JH, Craft J, Al Lozi R, Vaidya M, Meng Y, Deasy JO, et al. A Bayesian network approach for modeling local failure in lung cancer. Phys Med Biol. 2011; 56:1635–51.
Article23. Penson A, Camacho N, Zheng Y, Varghese AM, Al-Ahmadie H, Razavi P, et al. Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol. 2020; 6:84–91.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current status of genomic prediction using Multi-omics data in livestock
- Prediction of the Exposure to 1763MHz Radiofrequency Radiation Based on Gene Expression Patterns
- Integrating Deep Learning–Based Dose Distribution Prediction with Bayesian Networks for Decision Support in Radiotherapy for Upper Gastrointestinal Cancer
- Machine Learning vs. Statistical Model for Prediction Modelling: Application in Medical Imaging Research
- Development and Validation of Radiation Sensitivity Prediction Model Using Gene Expression Profiling Data Based on US National Cancer Institute-60 Tumor Cell Line