Ann Hepatobiliary Pancreat Surg.
2022 Feb;26(1):58-68. 10.14701/ahbps.21-108.
Identification of key genes and carcinogenic pathways in hepatitis B virus-associated hepatocellular carcinoma through bioinformatics analysis
- Affiliations
-
- 1Division of Liver Transplantation and Hepatobiliary Surgery, Department of Surgery, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 2Department of Surgery, Jeonbuk National University Hospital, Jeonju, Korea
- KMID: 2526833
- DOI: http://doi.org/10.14701/ahbps.21-108
Abstract
- Backgrounds/Aims
Mechanisms for the development of hepatocellular carcinoma (HCC) in hepatitis B virus (HBV)-infected patients remain unclear. The aim of the present study was to identify genes and pathways involved in the development of HBV-associated HCC.
Methods
The GSE121248 gene dataset, which included 70 HCCs and 37 adjacent liver tissues, was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) in HCCs and adjacent liver tissues were identified. Gene ontology and Kyoto Encyclopedia of Genes and Genome pathway enrichment analyses were then performed.
Results
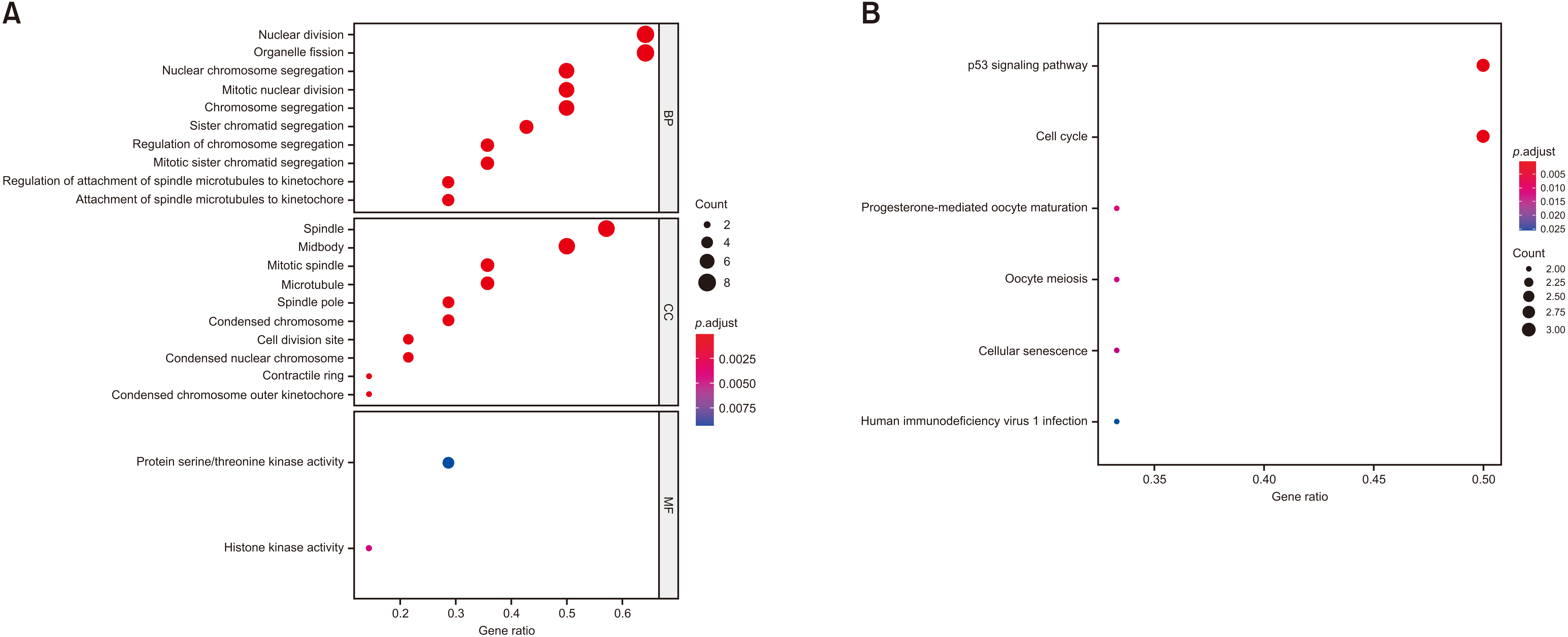
Of 134 DEGs identified, 34 were up-regulated and 100 were down-regulated in HCCs. The 34 up-regulated DEGs were mainly involved in nuclear division, organelle fission, spindle and midbody formation, histone kinase activity, and p53 signaling pathway, whereas the 100 down-regulated DEGs were involved in steroid and hormone metabolism, collagen-coated extracellular matrix, oxidoreductase activity, and activity on paired donors, including incorporation or reduction of molecular oxygen, monooxygenase activity, and retinol metabolism. Analyses of protein-protein interaction networks with a high degree of connectivity identified significant modules containing 14 hub genes, including ANLN , ASPM , BUB1B , CCNB1, CDK1, CDKN3, ECT2 , HMMR , NEK2 , PBK , PRC1, RACGAP1, RRM2 , and TOP2A , which were mainly associated with nuclear division, organelle fission, spindle formation, protein serine/threonine kinase activity, p53 signaling pathway, and cell cycle.
Conclusions
This study identified key genes and carcinogenic pathways that play essential roles in the development of HBV-associated HCC. This may provide important information for the development of diagnostic and therapeutic targets for HCC.
Keyword
Figure
Reference
-
1. Singal AG, Lampertico P, Nahon P. 2020; Epidemiology and surveillance for hepatocellular carcinoma: new trends. J Hepatol. 72:250–261. DOI: 10.1016/j.jhep.2019.08.025. PMID: 31954490. PMCID: PMC6986771.
Article2. Liu S, Yao X, Zhang D, Sheng J, Wen X, Wang Q, et al. 2018; Analysis of transcription factor-related regulatory networks based on bioinformatics analysis and validation in hepatocellular carcinoma. Biomed Res Int. 2018:1431396. DOI: 10.1155/2018/1431396. PMID: 30228980. PMCID: PMC6136478.
Article3. Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L. 2019; Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. J Cell Biochem. 120:10069–10081. DOI: 10.1002/jcb.28290. PMID: 30525236.
Article4. Forner A, Llovet JM, Bruix J. 2012; Hepatocellular carcinoma. Lancet. 379:1245–1255. DOI: 10.1016/S0140-6736(11)61347-0. PMID: 29307467.
Article5. Zhang L, Maddox AS. 2010; Anillin. Curr Biol. 20:R135–R136. DOI: 10.1016/j.cub.2009.12.017. PMID: 20178751. PMCID: PMC7176533.
Article6. Chuang HY, Ou YH. Abstract 4068: overexpression of anillin in colorectal cancer promotes the cell proliferation, cell mobility and cell invasion. Cancer Res. 2014; 74(19 Suppl):4068. DOI: 10.1158/1538-7445.AM2014-4068.
Article7. Zhang LH, Wang D, Li Z, Wang G, Chen DB, Cheng Q, et al. 2021; Overexpression of anillin is related to poor prognosis in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 20:337–344. DOI: 10.1016/j.hbpd.2020.08.007. PMID: 32933876.
Article8. Xu Z, Zhang Q, Luh F, Jin B, Liu X. 2019; Overexpression of the ASPM gene is associated with aggressiveness and poor outcome in bladder cancer. Oncol Lett. 17:1865–1876. DOI: 10.3892/ol.2018.9762. PMID: 30675249. PMCID: PMC6341836.
Article9. Xie JJ, Zhuo YJ, Zheng Y, Mo RJ, Liu ZZ, Li BW, et al. 2017; High expression of ASPM correlates with tumor progression and predicts poor outcome in patients with prostate cancer. Int Urol Nephrol. 49:817–823. DOI: 10.1007/s11255-017-1545-7. PMID: 28213802.
Article10. Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC, Peng SY, et al. 2008; ASPM is a novel marker for vascular invasion, early recurrence, and poor prognosis of hepatocellular carcinoma. Clin Cancer Res. 14:4814–4820. DOI: 10.1158/1078-0432.CCR-07-5262. PMID: 18676753.
Article11. Qiu J, Zhang S, Wang P, Wang H, Sha B, Peng H, et al. 2020; BUB1B promotes hepatocellular carcinoma progression via activation of the mTORC1 signaling pathway. Cancer Med. 9:8159–8172. DOI: 10.1002/cam4.3411. PMID: 32977361. PMCID: PMC7643650.
Article12. Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang M, et al. 2018; FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochem Biophys Res Commun. 500:924–929. DOI: 10.1016/j.bbrc.2018.04.201. PMID: 29705704.
Article13. Zhao J, Han SX, Ma JL, Ying X, Liu P, Li J, et al. 2013; The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 30:253–259. DOI: 10.3892/or.2013.2426. PMID: 23619525.
Article14. Zhou J, Han S, Qian W, Gu Y, Li X, Yang K. 2018; Metformin induces miR-378 to downregulate the CDK1, leading to suppression of cell proliferation in hepatocellular carcinoma. Onco Targets Ther. 11:4451–4459. DOI: 10.2147/OTT.S167614. PMID: 30104887. PMCID: PMC6074828.
Article15. Ito Y, Takeda T, Sakon M, Monden M, Tsujimoto M, Matsuura N. 2000; Expression and prognostic role of cyclin-dependent kinase 1 (cdc2) in hepatocellular carcinoma. Oncology. 59:68–74. DOI: 10.1159/000012140. PMID: 10895070.
Article16. Fan C, Chen L, Huang Q, Shen T, Welsh EA, Teer JK, et al. 2015; Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. Br J Cancer. 113:1735–1743. DOI: 10.1038/bjc.2015.378. PMID: 26554648. PMCID: PMC4701993.
Article17. Dai W, Miao H, Fang S, Fang T, Chen N, Li M. 2016; CDKN3 expression is negatively associated with pathological tumor stage and CDKN3 inhibition promotes cell survival in hepatocellular carcinoma. Mol Med Rep. 14:1509–1514. DOI: 10.3892/mmr.2016.5410. PMID: 27314282. PMCID: PMC4940071.
Article18. Chen J, Xia H, Zhang X, Karthik S, Pratap SV, Ooi LL, et al. 2015; ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol. 62:1287–1295. DOI: 10.1016/j.jhep.2015.01.014. PMID: 25617497.
Article19. Jin Y, Yu Y, Shao Q, Ma Y, Zhang R, Yao H, et al. 2014; Up-regulation of ECT2 is associated with poor prognosis in gastric cancer patients. Int J Clin Exp Pathol. 7:8724–8731. PMID: 25674238. PMCID: PMC4313974.20. He Z, Mei L, Connell M, Maxwell CA. 2020; Hyaluronan mediated motility receptor (HMMR) encodes an evolutionarily conserved homeostasis, mitosis, and meiosis regulator rather than a hyaluronan receptor. Cells. 9:819. DOI: 10.3390/cells9040819. PMID: 32231069. PMCID: PMC7226759.
Article21. Lin S, Zhou S, Jiang S, Liu X, Wang Y, Zheng X, et al. 2016; NEK2 regulates stem-like properties and predicts poor prognosis in hepatocellular carcinoma. Oncol Rep. 36:853–862. DOI: 10.3892/or.2016.4896. PMID: 27349376.
Article22. Fu L, Liu S, Wang H, Ma Y, Li L, He X, et al. 2017; Low expression of NEK2 is associated with hepatocellular carcinoma progression and poor prognosis. Cancer Biomark. 20:101–106. DOI: 10.3233/CBM-170586. PMID: 28759960.
Article23. Yang QX, Zhong S, He L, Jia XJ, Tang H, Cheng ST, et al. 2019; PBK overexpression promotes metastasis of hepatocellular carcinoma via activating ETV4-uPAR signaling pathway. Cancer Lett. 452:90–102. DOI: 10.1016/j.canlet.2019.03.028. PMID: 30914208.
Article24. Wang Y, Shi F, Xing GH, Xie P, Zhao N, Yin YF, et al. 2017; Protein regulator of cytokinesis PRC1 confers chemoresistance and predicts an unfavorable postoperative survival of hepatocellular carcinoma patients. J Cancer. 8:801–808. DOI: 10.7150/jca.17640. PMID: 28382142. PMCID: PMC5381168.
Article25. Imaoka H, Toiyama Y, Saigusa S, Kawamura M, Kawamoto A, Okugawa Y, et al. 2015; RacGAP1 expression, increasing tumor malignant potential, as a predictive biomarker for lymph node metastasis and poor prognosis in colorectal cancer. Carcinogenesis. 36:346–354. DOI: 10.1093/carcin/bgu327. PMID: 25568185.
Article26. Wang SM, Ooi LL, Hui KM. 2011; Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res. 17:6040–6051. DOI: 10.1158/1078-0432.CCR-11-0557. PMID: 21825042.
Article27. Nana AW, Wu SY, Yang YS, Chin YT, Cheng TM, Ho Y, et al. 2018; Nano-diamino-tetrac (NDAT) enhances resveratrol-induced antiproliferation by action on the RRM2 pathway in colorectal cancers. Horm Cancer. 9:349–360. DOI: 10.1007/s12672-018-0334-9. PMID: 30027502.
Article28. Lee B, Ha SY, Song DH, Lee HW, Cho SY, Park CK. 2014; High expression of ribonucleotide reductase subunit M2 correlates with poor prognosis of hepatocellular carcinoma. Gut Liver. 8:662–668. DOI: 10.5009/gnl13392. PMID: 25368754. PMCID: PMC4215454.
Article29. Zhao Q, Li H, Zhu L, Hu S, Xi X, Liu Y, et al. 2020; Bioinformatics analysis shows that TOP2A functions as a key candidate gene in the progression of cervical cancer. Biomed Rep. 13:21. DOI: 10.3892/br.2020.1328. PMID: 32765860. PMCID: PMC7403841.
Article30. Wong N, Yeo W, Wong WL, Wong NL, Chan KY, Mo FK, et al. 2009; TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance. Int J Cancer. 124:644–652. DOI: 10.1002/ijc.23968. PMID: 19003983.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A case of primary hepatocellular carcinoma following vertical transmission of hepatitis B virus in a child
- Serum Hepatitis B Virus DHA Level and Hepatocellulor Carcinoma
- The use of transient elastography for predicting hepatocellular carcinoma in chronic hepatitis B patients: Editorial on “Risk assessment of hepatitis B virus-related hepatocellular carcinoma development using vibration-controlled transient elastography: Systematic review and meta-analysis”
- Identification of multiple hub genes in acute kidney injury after kidney transplantation by bioinformatics analysis
- Relationships among Hepatitis C Virus, Hepatocellular Carcinoma, and Diffuse Large B Cell Lymphoma: A Case Report