Healthc Inform Res.
2022 Jan;28(1):46-57. 10.4258/hir.2022.28.1.46.
Texture, Morphology, and Statistical Analysis to Differentiate Primary Brain Tumors on Two-Dimensional Magnetic Resonance Imaging Scans Using Artificial Intelligence Techniques
- Affiliations
-
- 1Department of Computer Engineering, u-AHRC, Inje University, Gimhae, Korea
- 2Department of Digital Anti-Aging Healthcare, Inje University, Gimhae, Korea
- 3AI R&D Center, JLK Inc., Seoul, Korea
- KMID: 2526493
- DOI: http://doi.org/10.4258/hir.2022.28.1.46
Abstract
Objectives
A primary brain tumor starts to grow from brain cells, and it occurs as a result of errors in the DNA of normal cells. Therefore, this study was carried out to analyze the two-dimensional (2D) texture, morphology, and statistical features of brain tumors and to perform a classification using artificial intelligence (AI) techniques.
Methods
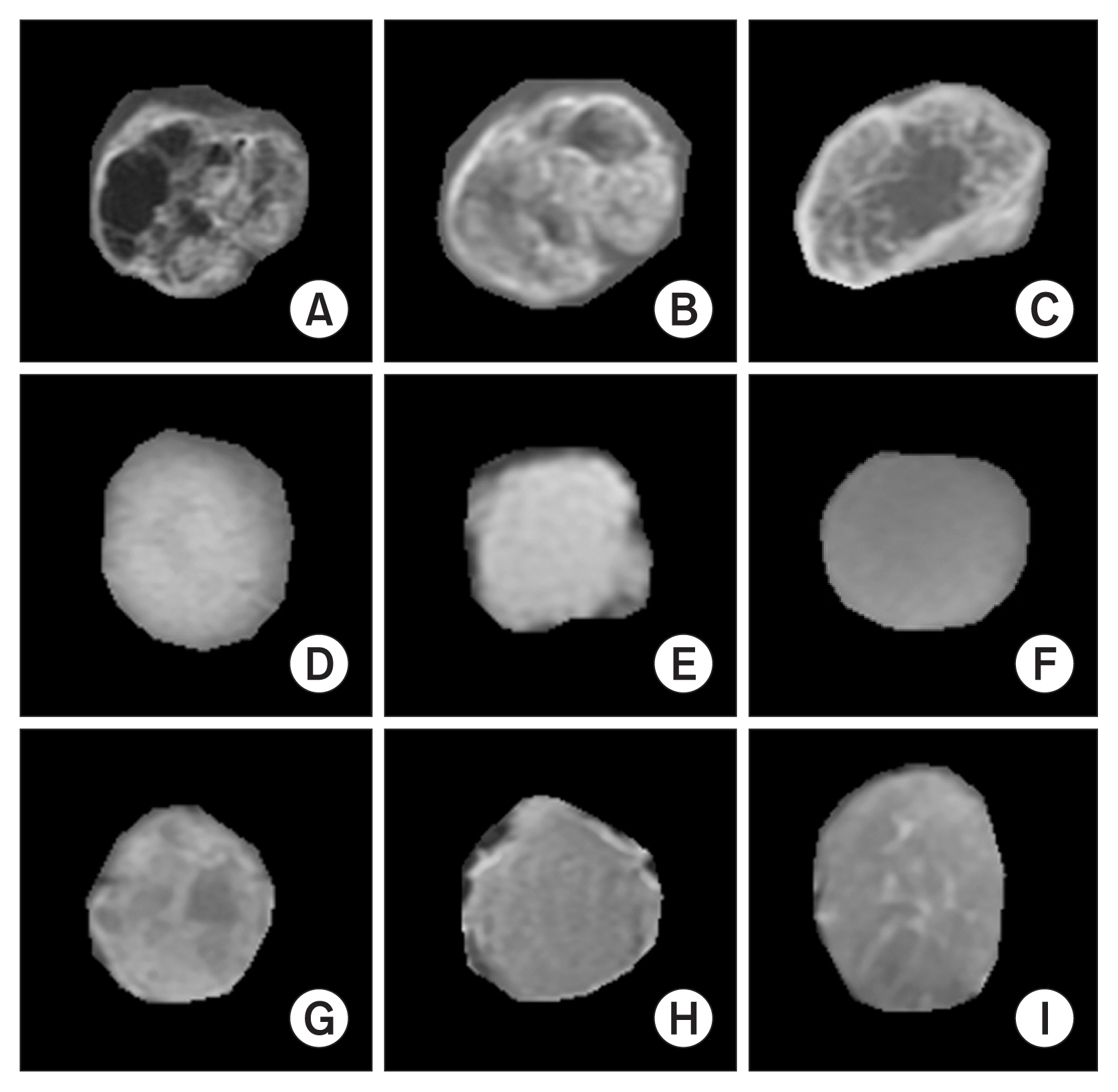
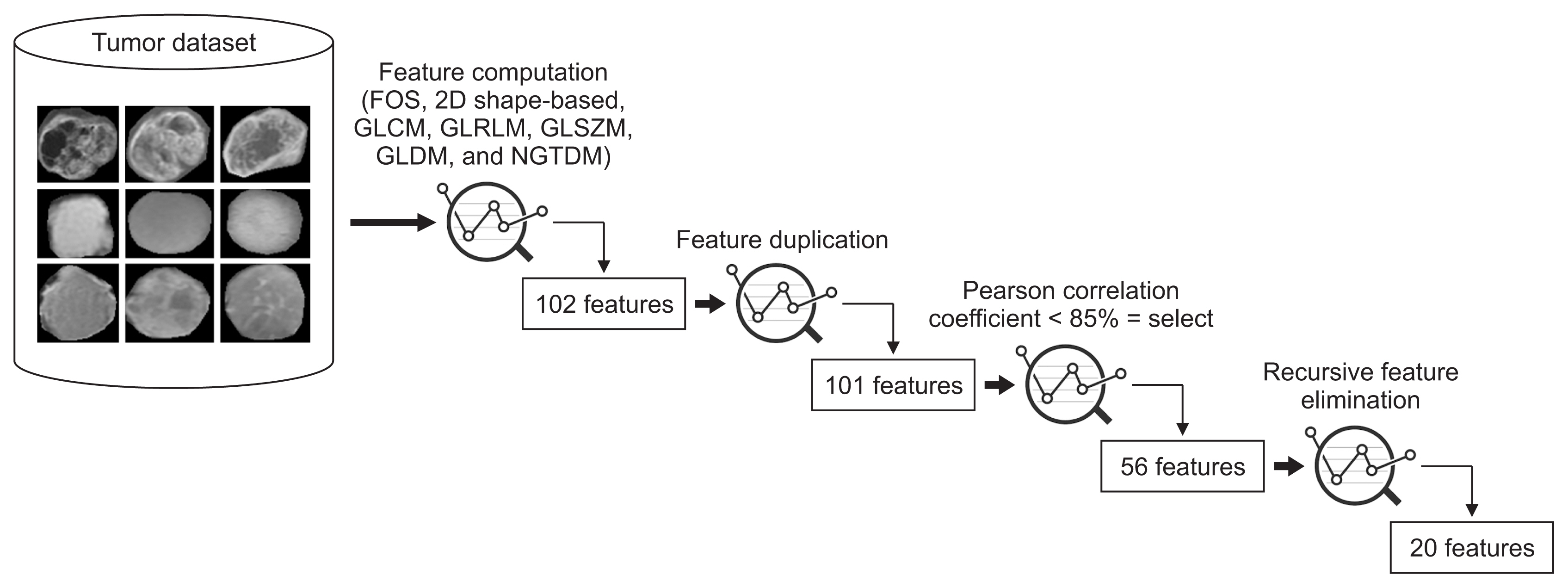
AI techniques can help radiologists to diagnose primary brain tumors without using any invasive measurement techniques. In this paper, we focused on deep learning (DL) and machine learning (ML) techniques for texture, morphological, and statistical feature classification of three tumor types (namely, glioma, meningioma, and pituitary). T1-weighted magnetic resonance imaging (MRI) 2D scans were used for analysis and classification (multiclass and binary). A total of 102 features were calculated for each tumor, and the 20 most significant features were selected using the three-step feature selection method, which included removing duplicate features, Pearson correlations, and recursive feature elimination.
Results
From the predicted results of multiclass and binary classification, a long short-term memory binary classification (glioma vs. meningioma) showed the best performance, with an average accuracy, recall, precision, F1-score, and kappa coefficient of 97.7%, 97.2%, 97.5%, 97.0%, and 94.7%, respectively.
Conclusions
The early diagnosis of primary brain tumors is very important because it can be the key to effective treatment. Therefore, this research presents a method for early diagnoses by effectively classifying three types of primary brain tumors.
Figure
Reference
-
References
1. Herholz K, Langen KJ, Schiepers C, Mountz JM. Brain tumors. Semin Nucl Med. 2012; 42(6):356–70.
Article2. Kheirollahi M, Dashti S, Khalaj Z, Nazemroaia F, Mahzouni P. Brain tumors: special characters for research and banking. Adv Biomed Res. 2015; 4:4.
Article3. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018; 392(10145):432–46.
Article4. Tunthanathip T, Kanjanapradit K, Ratanalert S, Phuenpathom N, Oearsakul T, Kaewborisutsakul A. Multiple, primary brain tumors with diverse origins and different localizations: case series and review of the literature. J Neurosci Rural Pract. 2018; 9(4):593–607.
Article5. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016; 131(6):803–20.
Article6. Escudero Sanchez L, Rundo L, Gill AB, Hoare M, Mendes Serrao E, Sala E. Robustness of radiomic features in CT images with different slice thickness, comparing liver tumour and muscle. Sci Rep. 2021; 11(1):8262.
Article7. Babu PR, Babu IR. ML and DL based classification model of lung cancer using nodule volume. In : Proceedings of 2020 4th International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud); 2020 Oct 7–9; Palladam, India. p. 1001–6.
Article8. Kim CH, Bhattacharjee S, Prakash D, Kang S, Cho NH, Kim HC, et al. Artificial intelligence techniques for prostate cancer detection through dual-channel tissue feature engineering. Cancers (Basel). 2021; 13(7):1524.
Article9. Cheng J. Brain tumor dataset [Internet]. London, UK: Figshare;2017. [cited at 2022 Jan 20]. Available from: https://figshare.com/articles/dataset/brain_tumor_data-set/1512427/5 .10. Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, et al. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS One. 2015; 10(10):e0140381.
Article11. Duron L, Balvay D, Vande Perre S, Bouchouicha A, Savatovsky J, Sadik JC, et al. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS One. 2019; 14(3):e0213459.
Article12. Chen CH, Chang CK, Tu CY, Liao WC, Wu BR, Chou KT, et al. Radiomic features analysis in computed tomography images of lung nodule classification. PLoS One. 2018; 13(2):e0192002.
Article13. Chaddad A. Automated feature extraction in brain tumor by magnetic resonance imaging using Gaussian mixture models. Int J Biomed Imaging. 2015; 2015:868031.
Article14. Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, et al. Shape and texture indexes application to cell nuclei classification. Int J Pattern Recognit Artif Intell. 2013; 27(01):1357002.
Article15. Wah YB, Ibrahim N, Hamid HA, Abdul-Rahman S, Fong S. Feature selection methods: case of filter and wrapper approaches for maximising classification accuracy. Pertanika J Sci Technol. 2018; 26(1):329–40.16. Hameed SS, Petinrin OO, Osman A, Hashi FS. Filterwrapper combination and embedded feature selection for gene expression data. Int J Adv Soft Comput Appl. 2018; 10(1):90–105.17. Hui KH, Ooi CS, Lim MH, Leong MS, Al-Obaidi SM. An improved wrapper-based feature selection method for machinery fault diagnosis. PLoS One. 2017; 12(12):e0189143.
Article18. Karim F, Majumdar S, Darabi H. Insights into LSTM fully convolutional networks for time series classification. IEEE Access. 2019; 7:67718–25.
Article19. Schuster M, Paliwal KK. Bidirectional recurrent neural networks. IEEE Trans Signal Process. 1997; 45(11):2673–81.
Article20. Ramachandran P, Zoph B, Le QV. Searching for activation functions. In : Proceedings of the 6th International Conference on Learning Representations (ICLR); 2018 Apr30–May 3; Vancouver, Canada.21. Sultan HH, Salem NM, Al-Atabany W. Multi-classification of brain tumor images using deep neural network. IEEE Access. 2019; 7:69215–25.
Article22. Machhale K, Nandpuru HB, Kapur V, Kosta L. MRI brain cancer classification using hybrid classifier (SVMKNN). In : Proceedings of 2015 International Conference on Industrial Instrumentation and Control (ICIC); 2015 May 28–30; Pune, India. p. 60–5.
Article23. Hossin M, Sulaiman MN. A review on evaluation metrics for data classification evaluations. Int J Data Min Knowl Manag Process. 2015; 5(2):1.24. Alqudah AM, Alquraan H, Qasmieh IA, Alqudah A, Al-Sharu W. Brain tumor classification using deep learning technique: a comparison between cropped, uncropped, and segmented lesion images with different sizes. Int J Adv Trends Comput Sci Eng. 2019; 8(6):3684–91.25. Pashaei A, Sajedi H, Jazayeri N. Brain tumor classification via convolutional neural network and extreme learning machines. In : Proceedings of 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE); 2018 Oct 25–26; Mashhad, Iran. p. 314–9.
Article26. Diaz-Pernas FJ, Martinez-Zarzuela M, Anton-Rodriguez M, Gonzalez-Ortega D. A Deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare (Basel). 2021; 9(2):153.27. Swati ZN, Zhao Q, Kabir M, Ali F, Ali Z, Ahmed S, et al. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph. 2019; 75:34–46.
Article28. Ismael MR, Abdel-Qader I. Brain tumor classification via statistical features and back-propagation neural network. In : Proceedings of 2018 IEEE International Conference on Electro/Information Technology (EIT); 2018 May 3–5; Rochester, MI. p. 252–7.
Article29. Badza MM, Barjaktarovic MC. Classification of brain tumors from MRI images using a convolutional neural network. Appl Sci. 2020; 10(6):1999.30. Cheng J, Yang W, Huang M, Huang W, Jiang J, Zhou Y, et al. Retrieval of brain tumors by adaptive spatial pooling and Fisher vector representation. PLoS One. 2016; 11(6):e0157112.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Advanced Magnetic Resonance Imaging for Pediatric Brain Tumors: Current Imaging Techniques and Interpretation Algorithms
- Applications of artificial intelligence in obstetrics
- Artificial Intelligence and Deep Learning in Musculoskeletal Magnetic Resonance Imaging
- Clinical Utilization of Brain Magnetic Resonance Imaging-Based Artificial Intelligence Software in the Spectrum of Alzheimer’s Disease: Case Series
- Artificial Intelligence in Neuroimaging: Clinical Applications