Clin Exp Otorhinolaryngol.
2021 Nov;14(4):399-406. 10.21053/ceo.2020.02124.
Genetic Variants and Clinical Phenotypes in Korean Patients With Hereditary Hemorrhagic Telangiectasia
- Affiliations
-
- 1Lee Gil Ya Cancer and Diabetes Institute, Gachon University, Incheon, Korea
- 2Department of Otolaryngology-Head and Neck Surgery, Gachon University Gil Medical Center, Incheon, Korea
- 3Department of Gastroenterology, Gachon University Gil Medical Center, Incheon, Korea
- 4Digestive Disease Center, CHA Bundang Medical Center, Seongnam, Korea
- 5Dr. Jin’s Premium Nose Clinic, Seoul, Korea
- 6Department of Otolaryngology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea
- 7Department of Otorhinolaryngology-Head and Neck Surgery, Kyung Hee University, School of Medicine, Seoul, Korea
- 8Department of Otorhinolaryngology-Head and Neck Surgery, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 9Department of Otorhinolaryngology-Head and Neck Surgery, Boramae Medical Center, Seoul National University College of Medicine, Seoul, Korea
- 10Department of Biochemistry, Gachon University College of Medicine, Incheon, Korea
- KMID: 2522431
- DOI: http://doi.org/10.21053/ceo.2020.02124
Abstract
Objectives
. Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant vascular disorder characterized by recurrent epistaxis, telangiectasia, and visceral arteriovenous malformations (AVMs). Activin A receptor-like type 1 (ACVRL1/ALK1) and endoglin (ENG) are the principal genes whose mutations cause HHT. No multicenter study has yet investigated correlations between genetic variations and clinical outcomes in Korean HHT patients.
Methods
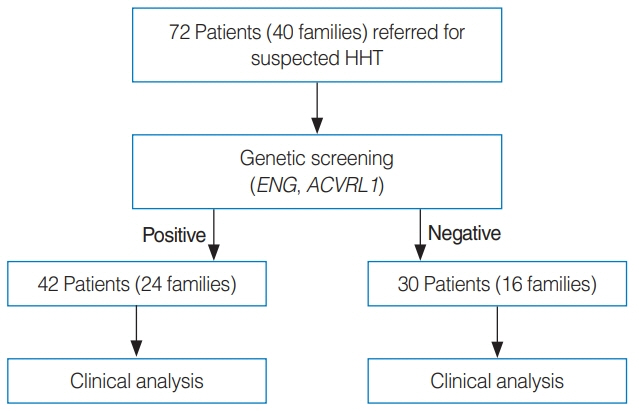
. Seventy-two members from 40 families suspected to have HHT based on symptoms were genetically screened for pathogenic variants of ACVRL1 and ENG. Patients with genetically diagnosed HHT were also evaluated.
Results
. In the HHT genetic screening, 42 patients from 24 of the 40 families had genetic variants that met the pathogenic criteria (pathogenic very strong, pathogenic strong, pathogenic moderate, or pathogenic supporting) based on the American College of Medical Genetics and Genomics Standards and Guidelines for either ENG or ACVRL1: 26 from 12 families (50%) for ENG, and 16 from 12 families (50%) for ACVRL1. Diagnostic screening of 42 genetically positive HHT patients based on the Curaçao criteria revealed that 24 patients (57%) were classified as having definite HHT, 17 (41%) as having probable HHT, and 1 (2%) as unlikely to have HHT. Epistaxis was the most common clinical presentation (38/42, 90%), followed by visceral AVMs (24/42, 57%) and telangiectasia (21/42, 50%). Five patients (12%) did not have a family history of HHT clinical symptoms.
Conclusion
. Only approximately half of patients with ACVRL1 or ENG genetic variants could be clinically diagnosed as having definite HHT, suggesting that genetic screening is important to confirm the diagnosis.
Figure
Reference
-
1. Shovlin CL. Hereditary haemorrhagic telangiectasia: pathophysiology, diagnosis and treatment. Blood Rev. 2010; Nov. 24(6):203–19.
Article2. Shovlin CL, Guttmacher AE, Buscarini E, Faughnan ME, Hyland RH, Westermann CJ, et al. Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu-Osler-Weber syndrome). Am J Med Genet. 2000; Mar. 91(1):66–7.
Article3. AAssar OS, Friedman CM, White RI Jr. The natural history of epistaxis in hereditary hemorrhagic telangiectasia. Laryngoscope. 1991; Sep. 101(9):977–80.
Article4. Kjeldsen AD, Kjeldsen J. Gastrointestinal bleeding in patients with hereditary hemorrhagic telangiectasia. Am J Gastroenterol. 2000; Feb. 95(2):415–8.5. Kjeldsen AD, Oxhoj H, Andersen PE, Green A, Vase P. Prevalence of pulmonary arteriovenous malformations (PAVMs) and occurrence of neurological symptoms in patients with hereditary haemorrhagic telangiectasia (HHT). J Intern Med. 2000; Sep. 248(3):255–62.
Article6. Larsen L, Marker CR, Kjeldsen AD, Poulsen FR. Prevalence of hereditary hemorrhagic telangiectasia in patients operated for cerebral abscess: a retrospective cohort analysis. Eur J Clin Microbiol Infect Dis. 2017; Oct. 36(10):1975–80.
Article7. McAllister KA, Grogg KM, Johnson DW, Gallione CJ, Baldwin MA, Jackson CE, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994; Dec. 8(4):345–51.8. Abdalla SA, Letarte M. Hereditary haemorrhagic telangiectasia: current views on genetics and mechanisms of disease. J Med Genet. 2006; Feb. 43(2):97–110.
Article9. McDonald J, Wooderchak-Donahue W, VanSant Webb C, Whitehead K, Stevenson DA, Bayrak-Toydemir P. Hereditary hemorrhagic telangiectasia: genetics and molecular diagnostics in a new era. Front Genet. 2015; Jan. 6:1.
Article10. University of Utah. ARUP scientific resource for research and education: HHT disease databases. Salt Lake City (UT): ARUP Laboratories;2011.11. Lee ST, Kim JA, Jang SY, Kim DK, Do YS, Suh GY, et al. Clinical features and mutations in the ENG, ACVRL1, and SMAD4 genes in Korean patients with hereditary hemorrhagic telangiectasia. J Korean Med Sci. 2009; Feb. 24(1):69–76.
Article12. Kim MJ, Kim ST, Lee HD, Lee KY, Seo J, Lee JB, et al. Clinical and genetic analyses of three Korean families with hereditary hemorrhagic telangiectasia. BMC Med Genet. 2011; Oct. 12:130.
Article13. Ha M, Kim YJ, Kwon KA, Hahm KB, Kim MJ, Kim DK, et al. Gastric angiodysplasia in a hereditary hemorrhagic telangiectasia type 2 patient. World J Gastroenterol. 2012; Apr. 18(15):1840–4.
Article14. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; Apr. 7(4):248–9.
Article15. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012; Jul. 40:W452–7.
Article16. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015; Aug. 31(16):2745–7.
Article17. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17(5):405–24.
Article18. Torring PM, Brusgaard K, Ousager LB, Andersen PE, Kjeldsen AD. National mutation study among Danish patients with hereditary haemorrhagic telangiectasia. Clin Genet. 2014; Aug. 86(2):123–33.
Article19. Chen YJ, Yang QH, Liu D, Liu QQ, Eyries M, Wen L, et al. Clinical and genetic characteristics of Chinese patients with hereditary haemorrhagic telangiectasia-associated pulmonary hypertension. Eur J Clin Invest. 2013; Oct. 43(10):1016–24.
Article20. Alaa El Din F, Patri S, Thoreau V, Rodriguez-Ballesteros M, Hamade E, Bailly S, et al. Functional and splicing defect analysis of 23 ACVRL1 mutations in a cohort of patients affected by hereditary hemorrhagic telangiectasia. PLoS One. 2015; Jul. 10(7):e0132111.
Article21. Komiyama M, Ishiguro T, Yamada O, Morisaki H, Morisaki T. Hereditary hemorrhagic telangiectasia in Japanese patients. J Hum Genet. 2014; Jan. 59(1):37–41.
Article22. Koenighofer M, Parzefall T, Frohne A, Allen M, Unterberger U, Laccone F, et al. Spectrum of novel hereditary hemorrhagic telangiectasia variants in an Austrian patient cohort. Clin Exp Otorhinolaryngol. 2019; Nov. 12(4):405–11.
Article23. Karlsson T, Cherif H. Mutations in the ENG, ACVRL1, and SMAD4 genes and clinical manifestations of hereditary haemorrhagic telangiectasia: experience from the Center for Osler’s Disease, Uppsala University Hospital. Ups J Med Sci. 2018; Sep. 123(3):153–7.
Article24. van Gent MW, Velthuis S, Post MC, Snijder RJ, Westermann CJ, Letteboer TG, et al. Hereditary hemorrhagic telangiectasia: how accurate are the clinical criteria. Am J Med Genet A. 2013; Mar. 161(3):461–6.
Article25. Wooderchak-Donahue WL, McDonald J, Farrell A, Akay G, Velinder M, Johnson P, et al. Genome sequencing reveals a deep intronic splicing ACVRL1 mutation hotspot in hereditary haemorrhagic telangiectasia. J Med Genet. 2018; Dec. 55(12):824–30.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Genetic Mutation Analysis Can Supplement Clinically Confirmed Hereditary Hemorrhagic Telangiectasia Populations
- Hereditary Hemorrhagic Telangiectasia in a Family
- Hereditary Hemorrhagic Telangiectasia Combined with Pulmonary Arteriovenous Malformation Treated with Transcatheter Embolotherapy
- A Case of Hereditary Hemorrhagic Telangiectasia
- Hepatic Involvement in Hereditary Hemorrhagic Telangiectasia