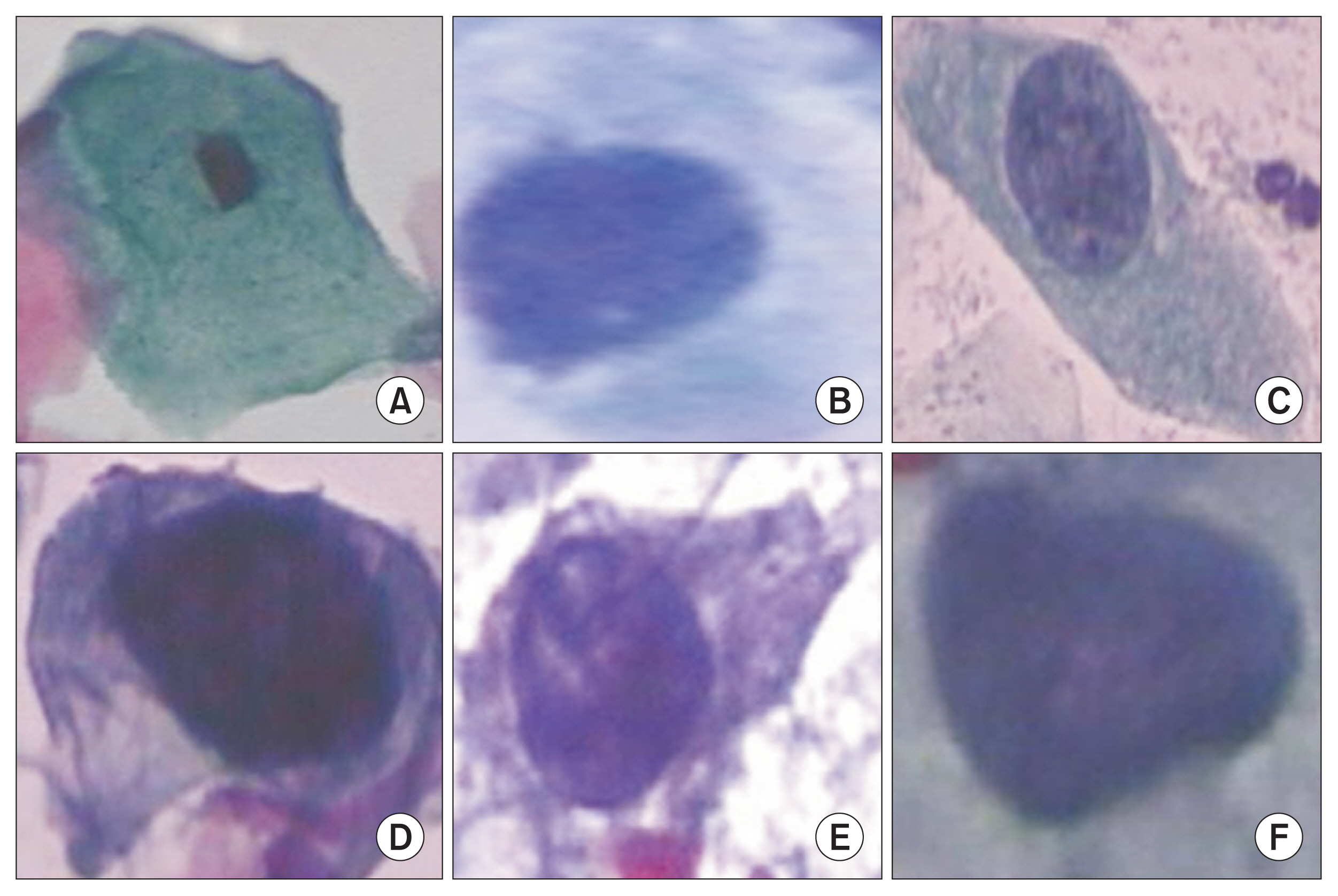
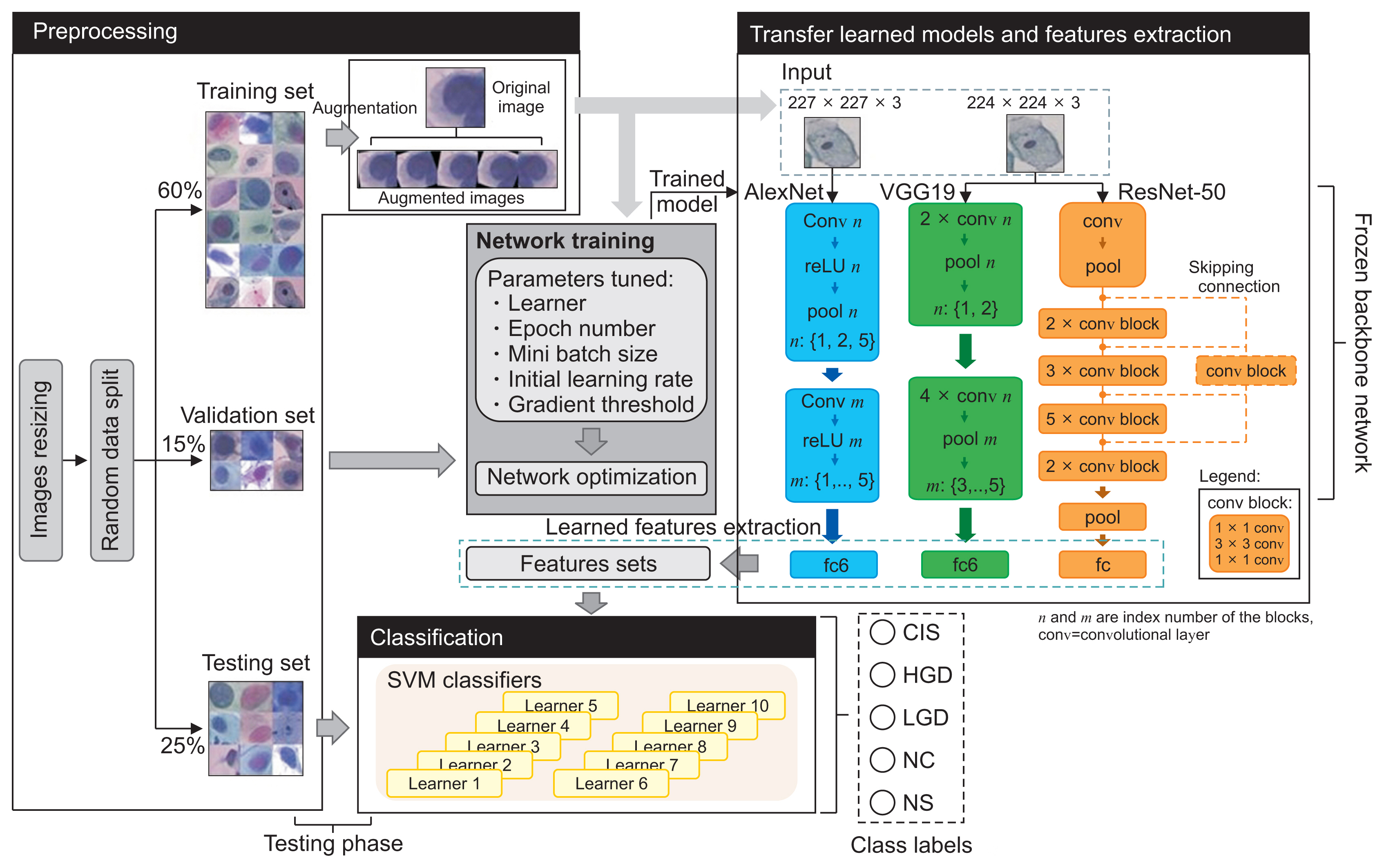
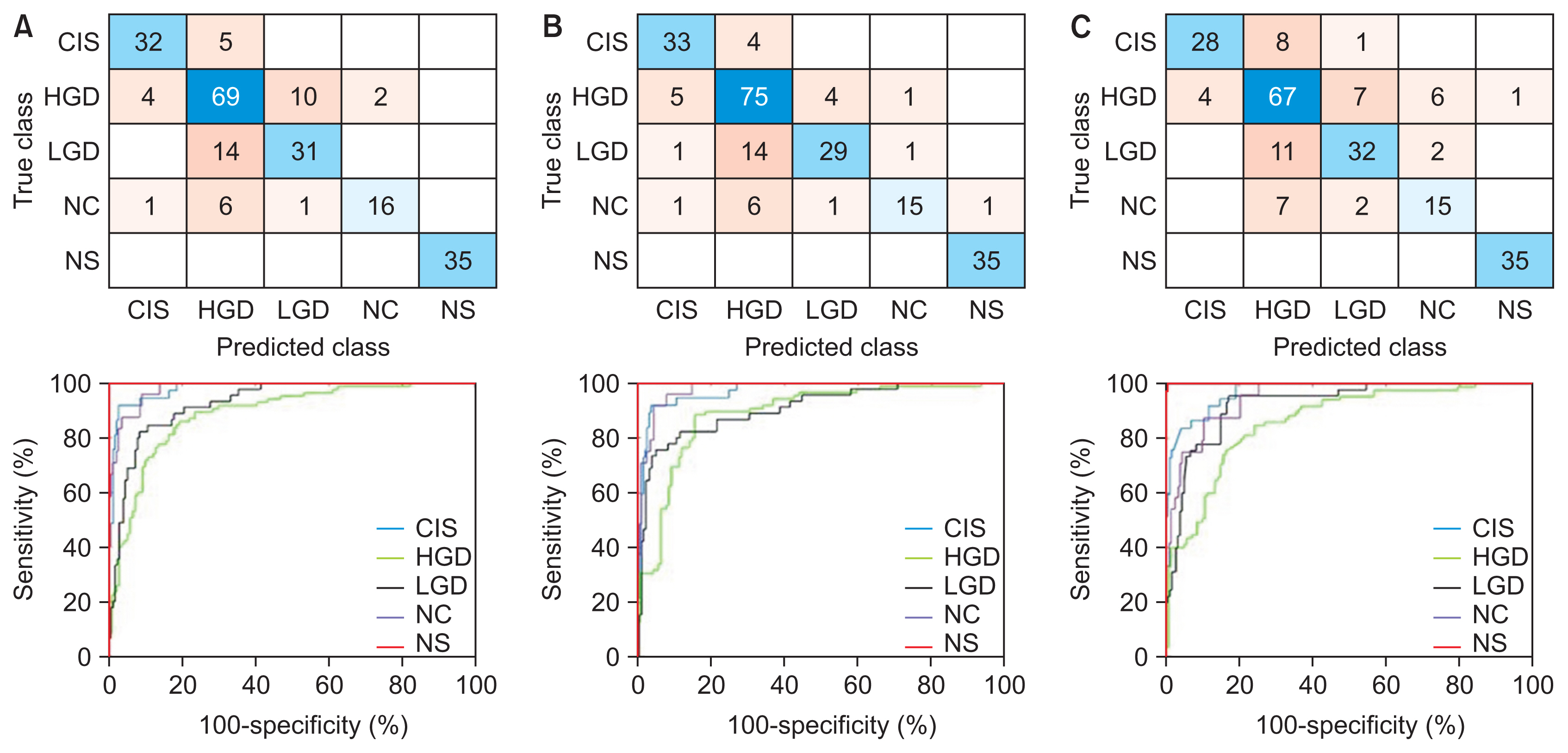
Healthc Inform Res.
2021 Oct;27(4):298-306. 10.4258/hir.2021.27.4.298.
Five-Class Classification of Cervical Pap Smear Images: A Study of CNN-Error-Correcting SVM Models
- Affiliations
-
- 1Faculty of Electrical and Electronic Engineering, Universiti Tun Hussein Onn Malaysia, Batu Pahat, Malaysia
- KMID: 2522211
- DOI: http://doi.org/10.4258/hir.2021.27.4.298
Abstract
Objectives
Different complex strategies of fusing handcrafted descriptors and features from convolutional neural network (CNN) models have been studied, mainly for two-class Papanicolaou (Pap) smear image classification. This paper explores a simplified system using combined binary coding for a five-class version of this problem.
Methods
This system extracted features from transfer learning of AlexNet, VGG19, and ResNet50 networks before reducing this problem into multiple binary sub-problems using error-correcting coding. The learners were trained using the support vector machine (SVM) method. The outputs of these classifiers were combined and compared to the true class codes for the final prediction.
Results
Despite the superior performance of VGG19-SVM, with mean ± standard deviation accuracy and sensitivity of 80.68% ± 2.00% and 80.86% ± 0.45%, respectively, this model required a long training time. There were also false-negative cases using both the VGGNet-SVM and ResNet-SVM models. AlexNet-SVM was more efficient in terms of running speed and prediction consistency. Our findings also showed good diagnostic ability, with an area under the curve of approximately 0.95. Further investigation also showed good agreement between our research outcomes and that of the state-of-the-art methods, with specificity ranging from 93% to 100%.
Conclusions
We believe that the AlexNet-SVM model can be conveniently applied for clinical use. Further research could include the implementation of an optimization algorithm for hyperparameter tuning, as well as an appropriate selection of experimental design to improve the efficiency of Pap smear image classification.
Figure
Reference
-
References
1. Pelkofski E, Stine J, Wages NA, Gehrig PA, Kim KH, Cantrell LA. Cervical cancer in women aged 35 years and younger. Clin Ther. 2016; 38(3):459–66.
Article2. Yoruk S, Acikgoz A, Ergor G. Determination of knowledge levels, attitude and behaviors of female university students concerning cervical cancer, human papiloma virus and its vaccine. BMC Womens Health. 2016; 16:51.
Article3. Watson M, Benard V, King J, Crawford A, Saraiya M. National assessment of HPV and Pap tests: changes in cervical cancer screening, National Health Interview Survey. Prev Med. 2017; 100:243–7.
Article4. MacLaughlin KL, Jacobson RM, Radecki Breitkopf C, Wilson PM, Jacobson DJ, Fan C, et al. Trends over time in pap and pap-HPV cotesting for cervical cancer screening. J Womens Health (Larchmt). 2019; 28(2):244–9.
Article5. Akbarzadeh MA, Hosseini MS. Implications for cancer care in Iran during COVID-19 pandemic. Radiother Oncol. 2020; 148:211–2.
Article6. Tsang-Wright F, Tasoulis MK, Roche N, MacNeill F. Breast cancer surgery after the COVID-19 pandemic. Future Oncol. 2020; 16(33):2687–90.
Article7. Price GJ, McCluggage WG, Morrison ML, McClean G, Venkatraman L, Diamond J, et al. Computerized diagnostic decision support system for the classification of preinvasive cervical squamous lesions. Hum Pathol. 2003; 34(11):1193–203.
Article8. Garrison A, Fischer AH, Karam AR, Leary A, Pieters RS. Cervical cancer. In : Pieters R, Rosenfeld J, Chen A, editors. Cancer concepts: a guidebook for the non-oncologist. Worcester (MA): University of Massachusetts Medical School;2015.9. William W, Ware A, Basaza-Ejiri AH, Obungoloch J. A review of image analysis and machine learning techniques for automated cervical cancer screening from pap-smear images. Comput Methods Programs Biomed. 2018; 164:15–22.
Article10. Selvathi D, Sharmila WR, Sankari PS. Advanced computational intelligence techniques based computer aided diagnosis system for cervical cancer detection using pap smear images. In : Dey N, Ashour AS, Borra S, editors. Classification in BioApps. Cham, Switzerland: Springer;2008. p. 295–322.11. Win KY, Choomchuay S, Hamamoto K, Raveesunthornkiat M, Rangsirattanakul L, Pongsawat S. Computer aided diagnosis system for detection of cancer cells on cytological pleural effusion images. Biomed Res Int. 2018; 2018:6456724.
Article12. Fekri-Ershad S. Pap smear classification using combination of global significant value, texture statistical features and time series features. Multimed Tools Appl. 2019; 78(22):31121–36.
Article13. Cervantes J, Garcia-Lamont F, Rodríguez-Mazahua L, Lopez A. A comprehensive survey on support vector machine classification: applications, challenges and trends. Neurocomputing. 2020; 408:189–215.
Article14. Chauhan VK, Dahiya K, Sharma A. Problem formulations and solvers in linear SVM: a review. Artif Intell Rev. 2019; 52(2):803–55.
Article15. Aoyagi K, Wang H, Sudo H, Chiba A. Simple method to construct process maps for additive manufacturing using a support vector machine. Addit Manuf. 2019; 27:353–62.
Article16. Taha B, Dias J, Werghi N. Classification of cervicalcancer using pap-smear images: a convolutional neural network approach. In : Valdes Hernandez M, Gonzalez-Castro V, editors. Medical image understanding and analysis. Cham, Switzerland: Springer;2017. p. 261–72.17. Khamparia A, Gupta D, de Albuquerque VH, Sangaiah AK, Jhaveri RH. Internet of health things-driven deep learning system for detection and classification of cervical cells using transfer learning. J Supercomput. 2020; 76:8590–608.
Article18. Jia AD, Li BZ, Zhang CC. Detection of cervical cancer cells based on strong feature CNN-SVM network. Neurocomputing. 2020; 411:112–27.
Article19. Chen W, Li X, Gao L, Shen W. Improving computer-aided cervical cells classification using transfer learning based snapshot ensemble. Appl Sci. 2020; 10(20):7292.
Article20. Kurnianingsih , Allehaibi KH, Nugroho LE, Lazuardi L, Prabuwono AS, Mantoro T. Segmentation and classification of cervical cells using deep learning. IEEE Access. 2019; 7:116925–41.
Article21. Ghoneim A, Muhammad G, Hossain MS. Cervical cancer classification using convolutional neural networks and extreme learning machines. Future Gener Comput Syst. 2020; 102:643–49.
Article22. Zhang L, Le Lu, Nogues I, Summers RM, Liu S, Yao J. DeepPap: deep convolutional networks for cervical cell classification. IEEE J Biomed Health Inform. 2017; 21(6):1633–43.
Article23. Wang P, Wang J, Li Y, Li L, Zhang H. Adaptive pruning of transfer learned deep convolutional neural network for classification of cervical pap smear images. IEEE Access. 2020; 8:50674–83.
Article24. Hussain E, Mahanta LB, Das CR, Talukdar RK. A comprehensive study on the multi-class cervical cancer diagnostic prediction on pap smear images using a fusion-based decision from ensemble deep convolutional neural network. Tissue Cell. 2020; 65:101347.
Article25. AlMubarak HA, Stanley J, Guo P, Long R, Antani S, Thoma G, et al. A hybrid deep learning and handcrafted feature approach for cervical cancer digital histology image classification. Int J Healthc Inf Syst Inform. 2019; 14(2):66–87.
Article26. Johnson DB, Rowlands CJ. Diagnosis and treatment of cervical intraepithelial neoplasia in general practice. BMJ. 1989; 299(6707):1083–6.
Article27. Mutombo AB, Simoens C, Tozin R, Bogers J, Van Geertruyden JP, Jacquemyn Y. Efficacy of commercially available biological agents for the topical treatment of cervical intraepithelial neoplasia: a systematic review. Syst Rev. 2019; 8(1):132.
Article28. Smith CM, Watson DI, Michael MZ, Hussey DJ. MicroRNAs, development of Barrett’s esophagus, and progression to esophageal adenocarcinoma. World J Gastroenterol. 2010; 16(5):531–7.
Article29. Pham DS, Venkatesh S. Joint learning and dictionary construction for pattern recognition. In : Proceedings of 2008 IEEE Conference on Computer Vision and Pattern Recognition; 2018 Jun 23–28; Anchorage, AK. p. 1–8.
Article30. Wang Q, Guo G. Benchmarking deep learning techniques for face recognition. J Vis Commun Image Represent. 2019; 65:102663.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Women's Practice and the Result of Pap Smear
- Correlation Between Cervicovaginal Cytology and Histopathological Study
- The comparison of Conventional Pap smears with Liquid Pap smears (PrepStain(TM) System) in Cervical Pap smears
- The Clinical Effectiveness of Cervicography in Cervical Cancer Screening
- Preliminary Report on Efficacy of Speculoscopy for Increasing the Sensitivity of Cervical Cancer Screening Test