Healthc Inform Res.
2021 Jan;27(1):39-47. 10.4258/hir.2021.27.1.39.
API Driven On-Demand Participant ID Pseudonymization in Heterogeneous Multi-Study Research
- Affiliations
-
- 1Department of Biomedical Informatics, University of Arkansas for Medical Sciences, Little Rock, AR, USA
- 2Department of Information Technology, University of Arkansas for Medical Sciences, Little Rock, AR, USA
- 3Department of Population Health Sciences, University of Texas Health Science Center at San Antonio, San Antonio, TX, USA
- KMID: 2512623
- DOI: http://doi.org/10.4258/hir.2021.27.1.39
Abstract
Objectives
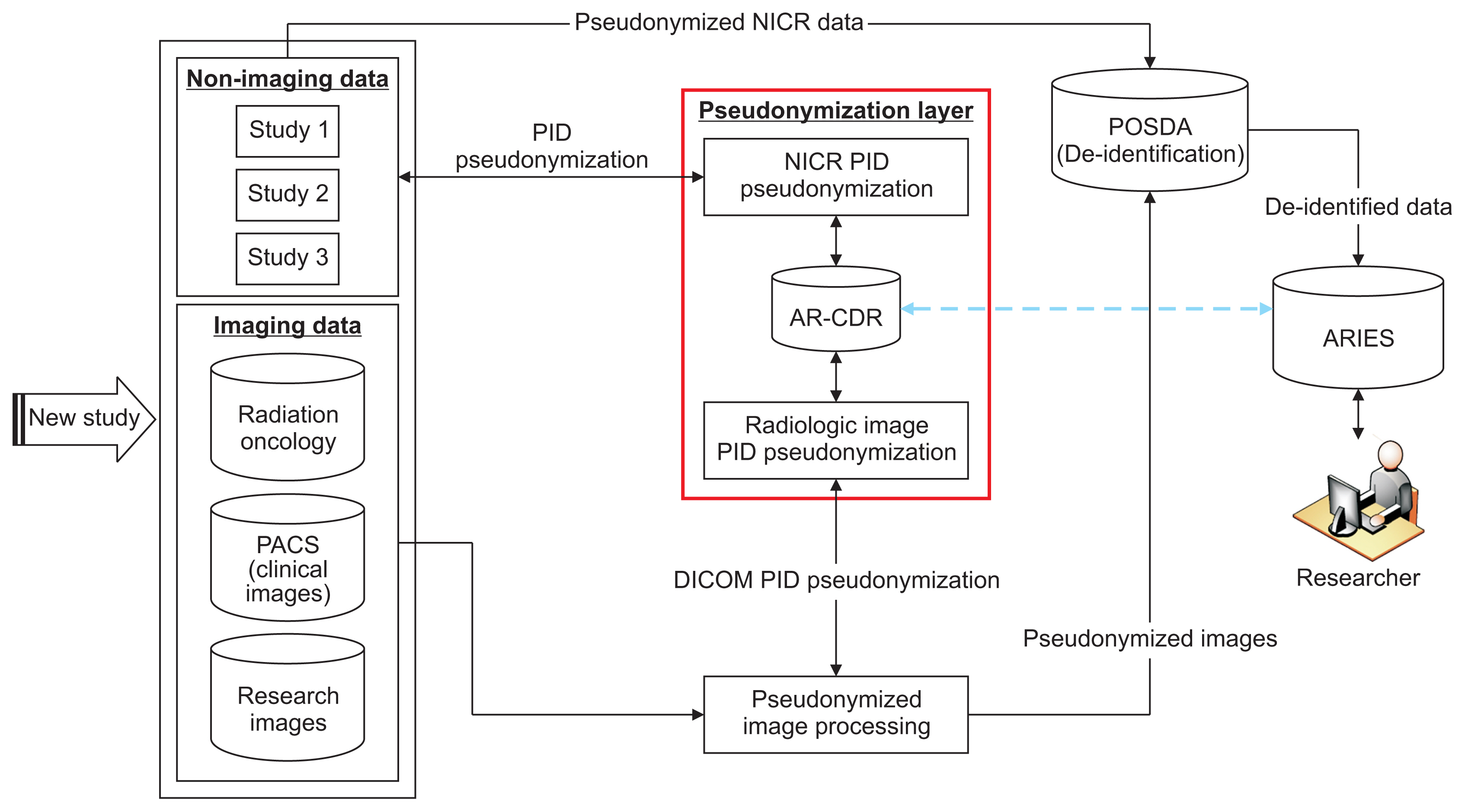
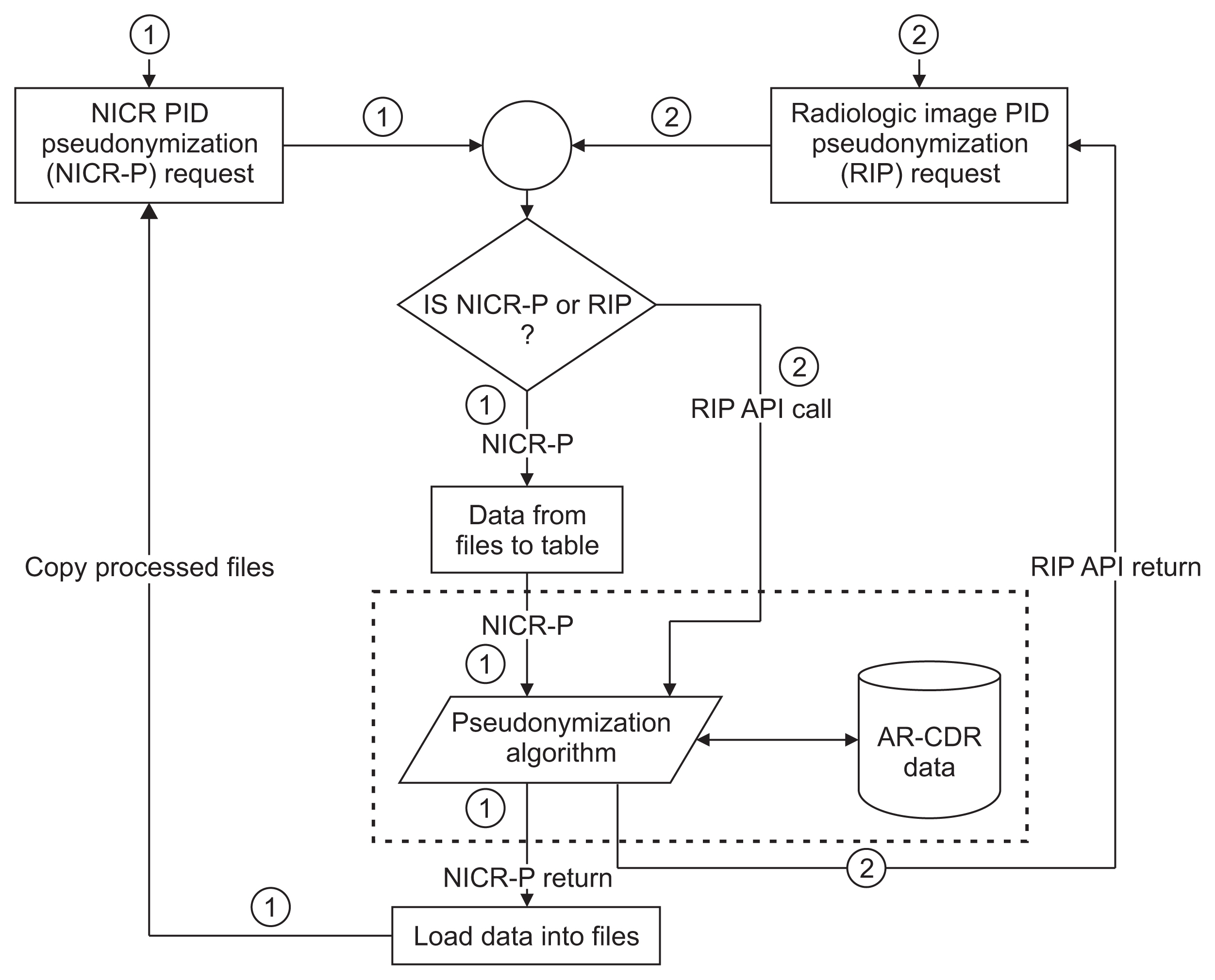
To facilitate clinical and translational research, imaging and non-imaging clinical data from multiple disparate systems must be aggregated for analysis. Study participant records from various sources are linked together and to patient records when possible to address research questions while ensuring patient privacy. This paper presents a novel tool that pseudonymizes participant identifiers (PIDs) using a researcher-driven automated process that takes advantage of application-programming interface (API) and the Perl Open-Source Digital Imaging and Communications in Medicine Archive (POSDA) to further de-identify PIDs. The tool, on-demand cohort and API participant identifier pseudonymization (O-CAPP), employs a pseudonymization method based on the type of incoming research data.
Methods
For images, pseudonymization of PIDs is done using API calls that receive PIDs present in Digital Imaging and Communications in Medicine (DICOM) headers and returns the pseudonymized identifiers. For non-imaging clinical research data, PIDs provided by study principal investigators (PIs) are pseudonymized using a nightly automated process. The pseudonymized PIDs (P-PIDs) along with other protected health information is further de-identified using POSDA.
Results
A sample of 250 PIDs pseudonymized by O-CAPP were selected and successfully validated. Of those, 125 PIDs that were pseudonymized by the nightly automated process were validated by multiple clinical trial investigators (CTIs). For the other 125, CTIs validated radiologic image pseudonymization by API request based on the provided PID and P-PID mappings.
Conclusions
We developed a novel approach of an ondemand pseudonymization process that will aide researchers in obtaining a comprehensive and holistic view of study participant data without compromising patient privacy.
Figure
Reference
-
References
1. Evans RS. Electronic Health Records: then, now, and in the future. Yearb Med Inform. 2016; Suppl 1(Suppl 1):S48–61.
Article2. Nordo AH, Levaux HP, Becnel LB, Galvez J, Rao P, Stem K, et al. Use of EHRs data for clinical research: historical progress and current applications. Learn Health Syst. 2019; 3(1):e10076.
Article3. Penning ML, Blach C, Walden A, Wang P, Donovan KM, Garza MY, et al. Near real time EHR data utilization in a clinical study. Stud Health Technol Inform. 2020; 270:337–41.4. Gliklich RE, Dreyer NA, Leavy MB. Registries for evaluating patient outcomes: a user’s guide. 3rd ed. Rockville (MD): Agency for Healthcare Research and Quality;2014.5. Nelson E, Talburt JR. Entity resolution for longitudinal studies in education using OYSTER. In : Proceedings of 2011 Information and Knowledge Engineering Conference (IKE); 2011 Jul 18–20; Las Vegas, NV. p. 286–90.6. Talburt JR, Zhou Y. A practical guide to entity resolution with OYSTER. Sadiq S, editor. Handbook of data quality. Heidelberg, Germany: Springer;2013. p. 235–70.
Article7. Erickson BJ, Buckner JC. Imaging in clinical trials. Cancer Inform. 2007; 4:13–8.
Article8. Grant JB, Hayes RP, Baker DW, Cangialose CB, Kieszak SM, Ballard DJ. Informatics, imaging, and healthcare quality management: imaging quality improvement opportunities and lessons learned form HCFA’s Health Care Quality Improvement Program. Clin Perform Qual Health Care. 1997; 5(3):133–9.9. Strickland NH. PACS (picture archiving and communication systems): filmless radiology. Arch Dis Child. 2000; 83(1):82–6.10. Digital Imaging and Communications in Medicine. DICOM standards [Internet]. Arlington (VA): DICOM;c2020. [cited 2020 Oct 23]. Available from: https://www.dicomstandard.org/current.11. Nass SJ, Levit LA, Gostin LO. Beyond the HIPAA Privacy Rule: enhancing privacy, improving health through research. Washington (DC): National Academies Press;2009.12. Linden T, Khandelwal R, Harkous H, Fawaz K. The privacy policy landscape after the GDPR. Proc Priv Enhanc Technol. 2020; (1):47–64.
Article13. Nelson GS. Practical implications of sharing data: a primer on data privacy, anonymization, and de-identification. In : Proceedings of SAS Global Forum; 2015 Apr 26–29; Dallas, TX. p. 1–23.14. Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. 2012; 50(Suppl):S82–101.
Article15. Chevrier R, Foufi V, Gaudet-Blavignac C, Robert A, Lovis C. Use and understanding of anonymization and de-identification in the biomedical literature: scoping review. J Med Internet Res. 2019; 21(5):e13484.
Article16. Kayaalp M. Modes of de-identification. AMIA Annu Symp Proc. 2018; 2017:1044–50.17. Riedl B, Neubauer T, Goluch G, Boehm O, Reinauer G, Krumboeck A. A secure architecture for the pseudonymization of medical data. In : Proceedings of the 2nd International Conference on Availability, Reliability and Security (ARES); 2007 Apr 10–13; Vienna, Austria. p. 318–24.
Article18. Aryanto KY, Oudkerk M, van Ooijen PM. Free DICOM de-identification tools in clinical research: functioning and safety of patient privacy. Eur Radiol. 2015; 25(12):3685–95.
Article19. Bennett W, Smith K, Jarosz Q, Nolan T, Bosch W. Reengineering workflow for curation of DICOM datasets. J Digit Imaging. 2018; 31(6):783–91.
Article20. Perl Open Source Digital Imaging and Communications in Medicine Archive (POSDA) [Internet] [place unknown]. github.com. 2019. [cited at 2020 Sep 17]. Available from: https://github.com/UAMS-DBMI/PosdaTools.21. Bruland P, Doods J, Brix T, Dugas M, Storck M. Connecting healthcare and clinical research: workflow optimizations through seamless integration of EHR, pseudonymization services and EDC systems. Int J Med Inform. 2018; 119:103–8.
Article22. Meystre SM, Lovis C, Burkle T, Tognola G, Budrionis A, Lehmann CU. Clinical data reuse or secondary use: current status and potential future progress. Yearb Med Inform. 2017; 26(1):38–52.
Article23. Fielding RT, Taylor RN. Architectural styles and the design of network-based software architectures. Irvine (CA): University of California;2000.24. Baghal A, Zozus M, Baghal A, Al-Shukri S, Prior F. Factors associated with increased adoption of a research data warehouse. Stud Health Technol Inform. 2019; 257:31–5.25. Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007; 25(11):1251–5.
Article26. Syed H, Talburt J, Liu F, Pullen D, Wu N. Developing and refining matching rules for entity resolution. In : Proceedings of the International Conference on Information and Knowledge Engineering (IKE); 2012 Jul 16–19; Las Vegas, NV.27. Foran DJ, Chen W, Chu H, Sadimin E, Loh D, Riedlinger G, et al. Roadmap to a comprehensive clinical data warehouse for precision medicine applications in oncology. Cancer Inform. 2017; 16:1176935117694349.
Article28. The Cancer Imaging Archive. Chest imaging with clinical and genomic correlates representing a rural COVID-19 positive population (COVID-19-AR) [Internet]. Fayetteville (AR): The Cancer Imaging Archive;2020. [cited at 2020 Sep 17]. Available from: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=70226443.29. Sood HS, Bates DW, Halamka JD, Sheikh A. Has the time come for a unique patient identifier for the U.S.? NEJM Catal. 2018; 4(1):1–4.30. Luthi S, Cohen JK. House votes to overturn ban on national patient identifier [Internet]. Chicago (IL): Modern Healthcare;2019. [cited at 2020 Oct 22]. Available from: https://www.modernhealthcare.com/politicspolicy/house-votes-overturn-ban-national-patientidentifier.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Microplate Identification System of Enterobacteriaceae
- Safe Utilization and Sharing of Genomic Data: Amendment to the Health and Medical Data Utilization Guidelines of South Korea
- Identification Results of Aerobic Gram-positive Bacteria Isolated from Blood Cultures Using BBL Crystal GP ID System
- Development of Simple Identification Method of Enterococci
- Data Pseudonymization in a Range That Does Not Affect Data Quality: Correlation with the Degree of Participation of Clinicians