Neonatal Med.
2020 Nov;27(4):207-213. 10.5385/nm.2020.27.4.207.
14q12q13.3 Deletion Diagnosed Using Chromosomal Microarray Analysis in an Infant Showing Seizures, Hypoplasia of the Corpus Callosum, and Developmental Delay
- Affiliations
-
- 1Departments of Pediatrics, Konyang University College of Medicine, Daejeon, Korea
- 2Departments of Medical Genetics, Konyang University College of Medicine, Daejeon, Korea
- KMID: 2510828
- DOI: http://doi.org/10.5385/nm.2020.27.4.207
Abstract
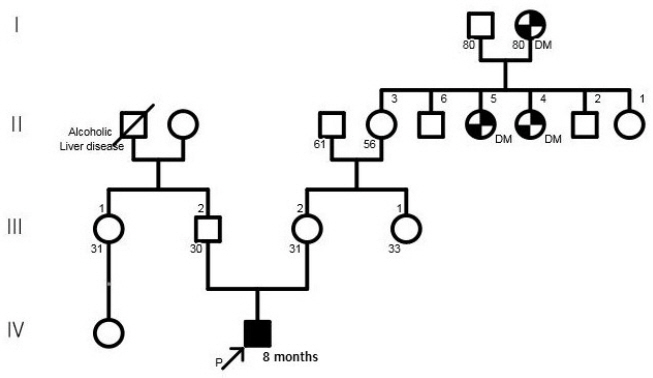
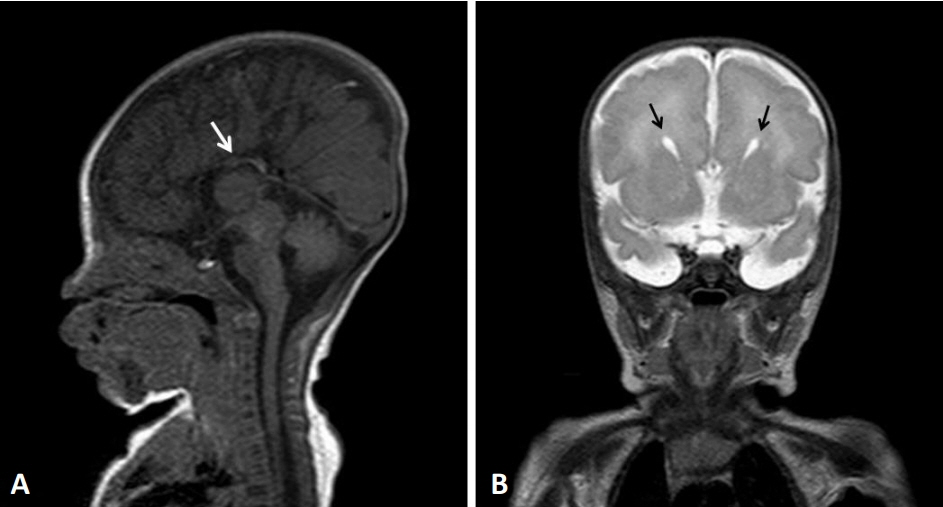
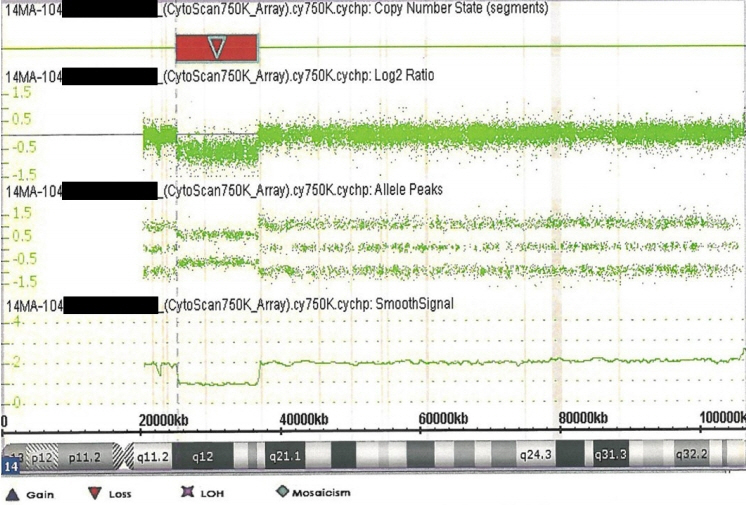
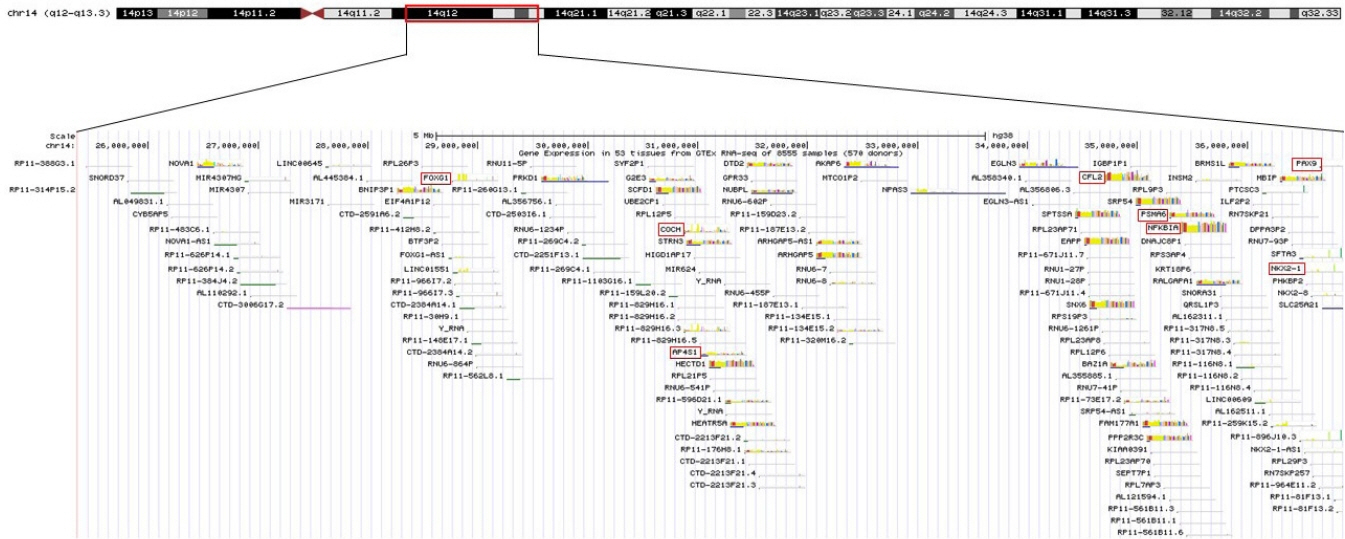
- 14q12q13.3 Deletion is a rare microdeletion syndrome associated with neurodevelopmental delay, failure to thrive, seizures, and abnormal brain development. Symptoms vary depending on the sites of gene deletion, and establishing the diagnosis is often difficult, as the condition cannot be detected with routine chromosome analysis. In this report, we present a patient with intrauterine growth retardation, microcephaly, muscle weakness, seizures, and hypoplasia of the corpus callosum who underwent diagnostic tests, including karyotyping in the neonatal period without leading to a specific diagnosis. The patient was confirmed with a serious developmental disorder, and a chromosomal microarray analysis was performed at 8 months of age, revealing a 14q12q13.3 deletion. In this case, the condition was diagnosed in early infancy, in contrast to previously reported cases, and the patient had diverse and severe symptoms. Establishing the diagnosis of 14q12q13.3 deletion syndrome allows better management of patient care and genetic counseling for the parents.
Keyword
Figure
Reference
-
1. Ponzi E, Gentile M, Agolini E, Matera E, Palumbi R, Buonadonna AL, et al. 14q12q13.2 microdeletion syndrome: clinical characterization of a new patient, review of the literature, and further evidence of a candidate region for CNS anomalies. Mol Genet Genomic Med. 2020; 8:e1289.2. Santen GW, Sun Y, Gijsbers AC, Carre A, Holvoet M, Haeringen Av, et al. Further delineation of the phenotype of chromosome 14q13 deletions: (positional) involvement of FOXG1 appears the main determinant of phenotype severity, with no evidence for a holoprosencephaly locus. J Med Genet. 2012; 49:366–72.3. Resta N, Memo L. Chromosomal microarray (CMA) analysis in infants with congenital anomalies: when is it really helpful? J Matern Fetal Neonatal Med. 2012; 25 Suppl 4:124–6.4. Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010; 86:749–64.5. Feldkamp ML, Carey JC, Byrne JL, Krikov S, Botto LD. Etiology and clinical presentation of birth defects: population based study. BMJ. 2017; 357:j2249.6. Brent RL. Addressing environmentally caused human birth defects. Pediatr Rev. 2001; 22:153–65.7. Mai CT, Isenburg JL, Canfield MA, Meyer RE, Correa A, Alverson CJ, et al. National population-based estimates for major birth defects, 2010-2014. Birth Defects Res. 2019; 111:1420–35.8. Hook EB. Contribution of chromosome abnormalities to human morbidity and mortality. Cytogenet Cell Genet. 1982; 33:101–6.9. Schinzel A. Catalog of unbalanced chromosome aberrations in man. 2nd ed. Berlin: Walter de Gruyter;2001.10. Emy Dorfman L, Leite JC, Giugliani R, Riegel M. Microarray-based comparative genomic hybridization analysis in neonates with congenital anomalies: detection of chromosomal imbalances. J Pediatr (Rio J). 2015; 91:59–67.11. Pickering DL, Eudy JD, Olney AH, Dave BJ, Golden D, Stevens J, et al. Array-based comparative genomic hybridization analysis of 1176 consecutive clinical genetics investigations. Genet Med. 2008; 10:262–6.12. Ellison JW, Ravnan JB, Rosenfeld JA, Morton SA, Neill NJ, Williams MS, et al. Clinical utility of chromosomal microarray analysis. Pediatrics. 2012; 130:e1085–95.13. Lalani SR. Current genetic testing tools in neonatal medicine. Pediatr Neonatol. 2017; 58:111–21.14. Florian C, Bahi-Buisson N, Bienvenu T. FOXG1-related disorders: from clinical description to molecular genetics. Mol Syndromol. 2012; 2:153–63.15. Kortum F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, Goldstein A, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011; 48:396–406.16. Hardies K, May P, Djemie T, Tarta-Arsene O, Deconinck T, Craiu D, et al. Recessive loss-of-function mutations in AP4S1 cause mild fever-sensitive seizures, developmental delay and spastic paraplegia through loss of AP-4 complex assembly. Hum Mol Genet. 2015; 24:2218–27.17. Gentile M, de Mattia D, Pansini A, Schettini F, Buonadonna AL, Capozza M, et al. 14q13 distal microdeletion encompassing NKX2-1 and PAX9: patient report and refinement of the associated phenotype. Am J Med Genet A. 2016; 170:1884–8.18. McDonald DR, Mooster JL, Reddy M, Bawle E, Secord E, Geha RS. Heterozygous N-terminal deletion of IkappaBalpha results in functional nuclear factor kappaB haploinsufficiency, ectodermal dysplasia, and immune deficiency. J Allergy Clin Immunol. 2007; 120:900–7.19. Fonseca DJ, Prada CF, Siza LM, Angel D, Gomez YM, Restrepo CM, et al. A de novo 14q12q13. 3 interstitial deletion in a patient affected by a severe neurodevelopmental disorder of unknown origin. Am J Med Genet A. 2012; 158A:689–93.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Case of 4q Deletion with Partial Agenesis of Corpus Callosum
- Diagnostic distal 16p11.2 deletion in a preterm infant with facial dysmorphism
- A Case of Chromosome 1q44 Deletion with Microcephaly and Multiple Congenital Anomalies
- A Case of Acrocallosal Syndrome with Developmental Delay: A case report
- Aicardi Syndrome: A case report