Intest Res.
2020 Oct;18(4):421-429. 10.5217/ir.2019.00120.
Pathophysiological role of Atg5 in human ulcerative colitis
- Affiliations
-
- 1Biochemistry Division, Department of Basic Sciences, School of Veterinary Medicine, Shiraz University, Shiraz, Iran
- 2Clinical Pathology Division, Department of Clinical Sciences, School of Veterinary Medicine, Shiraz University, Shiraz, Iran
- 3Health Policy Research Center, Shiraz University of Medical Sciences, Shiraz, Iran
- 4Cardiovascular Research Center, Shiraz University of Medical Sciences, Shiraz, Iran
- 5New Iberia Research Center, University of Louisiana at Lafayette, Lafayette, LA, USA
- KMID: 2508570
- DOI: http://doi.org/10.5217/ir.2019.00120
Abstract
- Background/Aims
Ulcerative colitis (UC), along with Crohn’s disease, is one of the main types of inflammatory bowel disease (IBD). On the other hand, deregulated autophagy is involved in many chronic diseases, including IBD. In this study, we aimed to investigate the role of Atg5 and microRNA-181a (miR-181a) in the pathophysiology of UC.
Methods
Colon biopsy, stool, and blood samples of 6 men and 9 women were confirmed for UC. Also, 13 men and 17 women were selected as healthy control (HC). Enzyme-linked immunosorbent assay (ELISA) and immunohistochemistry were used to measure the Atg-5 content of the colon biopsies. Besides, the serum and stool levels of Atg5 were measured using ELISA. Moreover, the total RNA of blood cells was extracted and evaluated for the expression of miR-181a.
Results
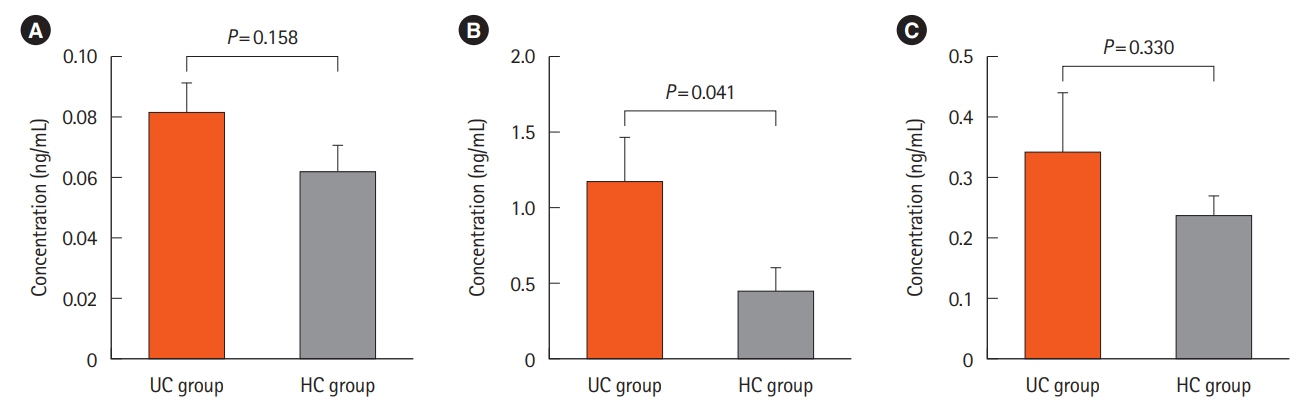
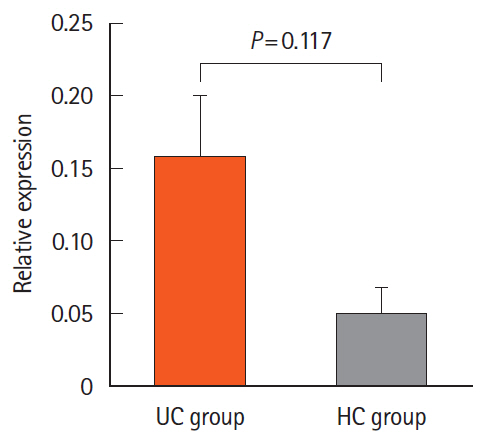
We found 1.2 ng/mL versus 0.46 ng/mL, 0.34 ng/mL versus 0.24 ng/mL, and 0.082 ng/mL versus 0.062 ng/mL of Atg5 in stool, intestinal tissue, and serum of UC and HCs, respectively. There was no significant difference in the expression of miR-181a in the blood samples of UC and HCs. Immunohistochemistry showed high positivity without any significant difference between the 2 groups in the quantitative analysis.
Conclusions
The significant difference observed between the stool Atg5 content of the HCs and UC patients may provide new insight into using this protein as a diagnostic biomarker, however, considering the small size of our studied population further studies are needed.
Keyword
Figure
Reference
-
1. Hosseini SV, Taghavi SA, Jafari P, et al. Incidence of ulcerative colitis relapse: a prospective cohort study in southern Iran. Ann Colorectal Res. 2016; 4:e34565.
Article2. Iida T, Onodera K, Nakase H. Role of autophagy in the pathogenesis of inflammatory bowel disease. World J Gastroenterol. 2017; 23:1944–1953.
Article3. Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. Am J Gastroenterol Suppl. 2012; 1:15.
Article4. Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011; 140:1785–1794.
Article5. Ramsey M, Krishna SG, Stanich PP, et al. Inflammatory bowel disease adversely impacts colorectal cancer surgery short-term outcomes and health-care resource utilization. Clin Transl Gastroenterol. 2017; 8:e127.
Article6. Diefenbach KA, Breuer CK. Pediatric inflammatory bowel disease. World J Gastroenterol. 2006; 12:3204–3212.
Article7. El-Khider F, McDonald C. Links of autophagy dysfunction to inflammatory bowel disease onset. Dig Dis. 2016; 34:27–34.
Article8. Yu L, Chen Y, Tooze SA. Autophagy pathway: cellular and molecular mechanisms. Autophagy. 2018; 14:207–215.
Article9. Simon HU, Friis R. Autophagy signalling. eLS. [published online ahead of print December 14, 2016]. https://doi.org/10.1002/9780470015902.a0026792.
Article10. Esclatine A, Chaumorcel M, Codogno P. Macroautophagy signaling and regulation. Curr Top Microbiol Immunol. 2009; 335:33–70.
Article11. Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007; 39:207–211.
Article12. Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007; 39:596–604.13. Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011; 60:1580–1588.
Article14. Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012; 45:629–641.
Article15. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011; 147:728–741.
Article16. Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012; 33:2018–2025.
Article17. Huang Y, Guerrero-Preston R, Ratovitski EA. PhosphoΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012; 11:1247–1259.
Article18. Jing Z, Han W, Sui X, Xie J, Pan H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015; 356(2 Pt B):332–338.
Article19. Lankarani KB, Sepehrimanesh M, Seghatoleslam SF, Hoseini SE, Ghavami S. Autophagy-related protein 7 level in patients with ulcerative colitis. Scand J Gastroenterol. 2017; 52:468.
Article20. Hao X, Yang B, Liu X, Yang H, Liu X. Expression of Beclin1 in the colonic mucosa tissues of patients with ulcerative colitis. Int J Clin Exp Med. 2015; 8:21098–21105.21. Paiva NM, Pascoal LB, Negreiros LMV, et al. Ileal pouch of ulcerative colitis and familial adenomatous polyposis patients exhibit modulation of autophagy markers. Sci Rep. 2018; 8:2619.
Article22. Zhao H, Xi H, Wei B, et al. Expression of decorin in intestinal tissues of mice with inflammatory bowel disease and its correlation with autophagy. Exp Ther Med. 2016; 12:3885–3892.
Article23. Elliott TR, Hudspith BN, Rayment NB, et al. Defective macrophage handling of Escherichia coli in Crohn’s disease. J Gastroenterol Hepatol. 2015; 30:1265–1274.
Article24. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012; 491:119–124.25. Cadwell K, Liu J, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008; 456:259–263.
Article26. Tekirdag KA, Korkmaz G, Ozturk DG, Agami R, Gozuacik D. MIR181A regulates starvation- and rapamycin-induced autophagy through targeting of ATG5. Autophagy. 2013; 9:374–385.
Article27. Chapman CG, Pekow J. The emerging role of miRNAs in inflammatory bowel disease: a review. Therap Adv Gastroenterol. 2015; 8:4–22.28. Li M, Zhang S, Qiu Y, et al. Upregulation of miR-665 promotes apoptosis and colitis in inflammatory bowel disease by repressing the endoplasmic reticulum stress components XBP1 and ORMDL3. Cell Death Dis. 2017; 8:e2699.
Article29. Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008; 135:1624–1635.
Article30. Wang S, Huang Y, Zhou C, et al. The role of autophagy and related microRNAs in inflammatory bowel disease. Gastroenterol Res Pract. 2018; 2018:7565076.
Article31. Cao B, Zhou X, Ma J, et al. Role of miRNAs in inflammatory bowel disease. Dig Dis Sci. 2017; 62:1426–1438.
Article32. Kalla R, Ventham NT, Kennedy NA, et al. MicroRNAs: new players in IBD. Gut. 2015; 64:504–517.
Article33. Pierdomenico M, Cesi V, Cucchiara S, et al. NOD2 is regulated by Mir-320 in physiological conditions but this control is altered in inflamed tissues of patients with inflammatory bowel disease. Inflamm Bowel Dis. 2016; 22:315–326.
Article34. Zhai Z, Wu F, Dong F, et al. Human autophagy gene ATG16L1 is post-transcriptionally regulated by MIR142-3p. Autophagy. 2014; 10:468–479.
Article35. Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014; 146:508–519.
Article36. Seoudi AM, Lashine YA, Abdelaziz AI. MicroRNA-181a: a tale of discrepancies. Expert Rev Mol Med. 2012; 14:e5.37. Xie W, Li M, Xu N, et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One. 2013; 8:e58639.
Article38. Cosin-Roger J, Simmen S, Melhem H, et al. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat Commun. 2017; 8:98.
Article39. Tang JY, Fang YY, Hsi E, et al. Immunopositivity of Beclin-1 and ATG5 as indicators of survival and disease recurrence in oral squamous cell carcinoma. Anticancer Res. 2013; 33:5611–5616.40. Cho DH, Jo YK, Kim SC, Park IJ, Kim JC. Down-regulated expression of ATG5 in colorectal cancer. Anticancer Res. 2012; 32:4091–4096.41. Wang SL, Shao BZ, Zhao SB, et al. Impact of Paneth cell autophagy on inflammatory bowel disease. Front Immunol. 2018; 9:693.
Article42. Randall-Demllo S, Chieppa M, Eri R. Intestinal epithelium and autophagy: partners in gut homeostasis. Front Immunol. 2013; 4:301.
Article43. Ke P, Shao BZ, Xu ZQ, Chen XW, Liu C. Intestinal autophagy and its pharmacological control in inflammatory bowel disease. Front Immunol. 2017; 7:695.
Article