Blood Res.
2020 Sep;55(3):131-138. 10.5045/br.2020.2020080.
Genomic alterations in chronic lymphocytic leukemia and their correlation with clinico-hematological parameters and disease progression
- Affiliations
-
- 1Departments of Hematology , Postgraduate Institute of Medical Education and Research, Chandigarh, India
- 2Departments of Internal Medicine, Postgraduate Institute of Medical Education and Research, Chandigarh, India
- KMID: 2507052
- DOI: http://doi.org/10.5045/br.2020.2020080
Abstract
- Background
Chronic lymphocytic leukemia (CLL) is a heterogeneous disease, which is attributed to differences in the genetic characteristics of the leukemic clone. We studied the genomic profile of 52 treatment-naïve CLL patients.
Methods
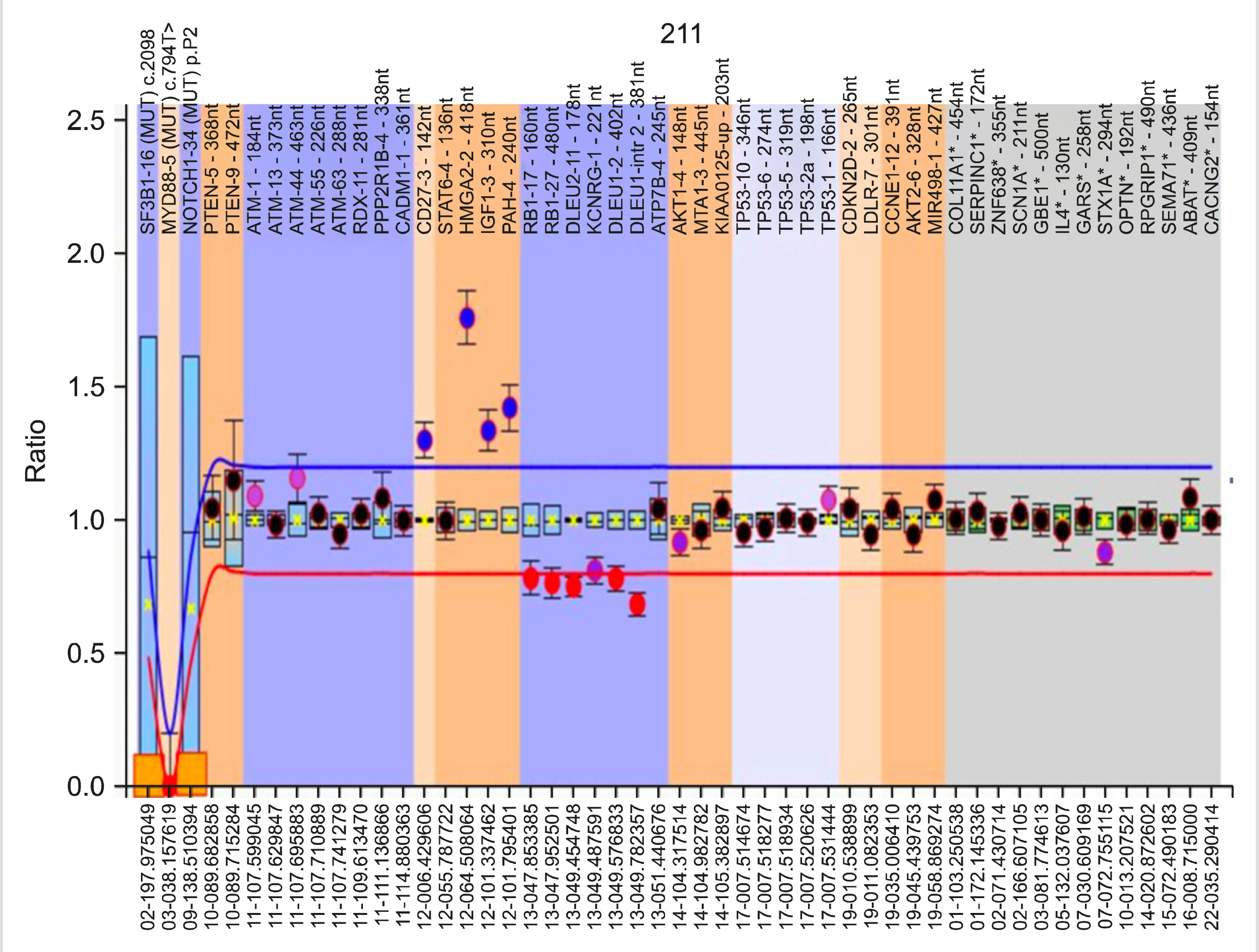
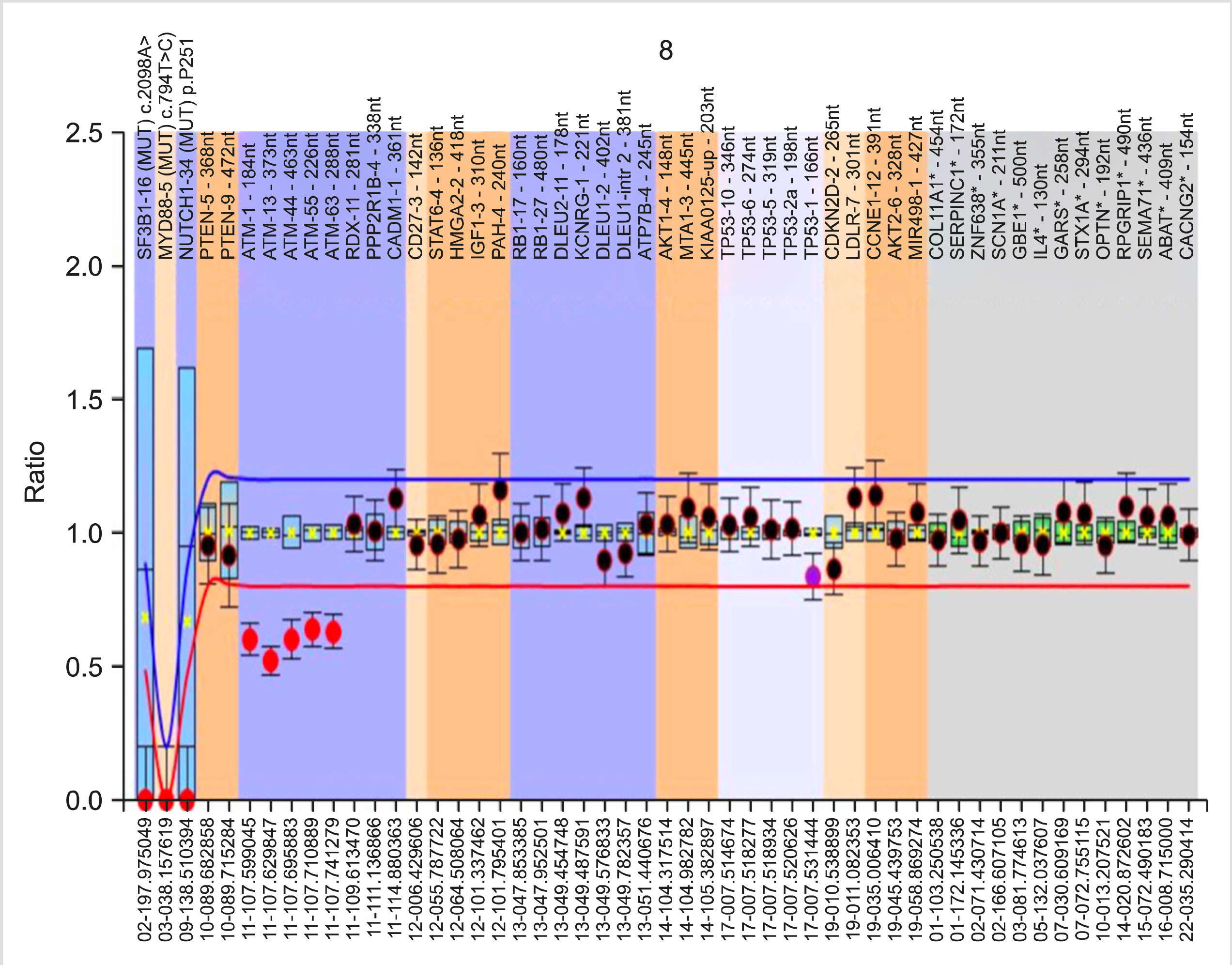
Genetic analysis was performed by multiplex ligation-dependent probe amplification (MLPA) using the SALSA P038 Probemix (MRC Holland, Amsterdam), which contains probes for 2p (MYCN,ALK,REL), 6q, 8p (TNFRSF10A/B), 8q (EIF3H,MYC), 9p21 (CDKN2A/B), 10q (PTEN), 11q (ATM, RDX, PPP2R1B, CADM1), chromosome 12, 13q14 (RB1, DLEU1/2/7, KCNRG, MIR15A), 14q, 17p (TP53) and chromosome 19, and for NOTCH1 7541-7542delCT, SF3B1 K700E, and MYD88 L265P mutations.
Results
The median age was 65 years (male:female=2:1). The median hemoglobin, total leukocyte, and platelet counts were 12.4 g/dL, 57.7×10 9 /L, and 176.5×10 9 /L, respectively. At least one genetic abnormality was observed in 34 (65%) patients. The most common abnormality was del(13q14) (deleted DLEU2 and DLEU1/RB1 genes), which was observed in 22 (42%) cases, followed by trisomy 12 [7 (13%) cases]. Del(11q) (deleted ATM, RDX11/PPP2R1B-4) and del(17p) (deleted TP53) were present in 5 (10%) and 2 (4%) cases, respectively. 19p13.2 (CDKN2D-2) amplification and NOTCH1 mutation were found in one case each.
Conclusion
Genetic abnormalities are commonly (65%) observed in CLL patients. Del(13q), which is associated with DLEU2 andDLEU1/RB1 gene deletion, was the most common. Compared with other abnormalities, del(11q) and del(17p) patients presented with cytopenia and higher Binet stage, while those with del(13q14) had a longer time to first treatment.
Keyword
Figure
Reference
-
1. Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. 2010; From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev. 10:37–50. DOI: 10.1038/nrc2764. PMID: 19956173.
Article2. Sutton LA, Rosenquist R. 2015; The complex interplay between cell-intrinsic and cell-extrinsic factors driving the evolution of chronic lymphocytic leukemia. Semin Cancer Biol. 34:22–35. DOI: 10.1016/j.semcancer.2015.04.009. PMID: 25963298.
Article3. Vollbrecht C, Mairinger FD, Koitzsch U, et al. 2015; Comprehensive analysis of disease-related genes in chronic lymphocytic leukemia by multiplex PCR-based next genera-tion sequencing. PLoS One. 10:e0129544. DOI: 10.1371/journal.pone.0129544. PMID: 26053404. PMCID: PMC4459702.
Article4. Döhner H, Stilgenbauer S, Benner A, et al. 2000; Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 343:1910–6. DOI: 10.1056/NEJM200012283432602. PMID: 11136261.
Article5. Ries LAG, Harkins D, Krapcho M, et al. 2007. SEER cancer statistics review, 1975-2004. National Cancer Institute;Bethesda, MD: at https://seer.cancer.gov/archive/csr/1975_2004/. Accessed March 2, 2020.6. Gunawardana C, Austen B, Powell JE, et al. 2008; South Asian chronic lymphocytic leukaemia patients have more rapid disease progression in comparison to White patients. Br J Haematol. 142:606–9. DOI: 10.1111/j.1365-2141.2008.07226.x. PMID: 18503582.
Article7. Hömig-Hölzel C, Savola S. 2012; Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 21:189–206. DOI: 10.1097/PDM.0b013e3182595516. PMID: 23111197.
Article8. Al Zaabi EA, Fernandez LA, Sadek IA, Riddell DC, Greer WL. 2010; Multiplex ligation-dependent probe amplification versus multiprobe fluorescence in situ hybridization to detect genomic aberrations in chronic lymphocytic leukemia: a tertiary center experi-ence. J Mol Diagn. 12:197–203. DOI: 10.2353/jmoldx.2010.090046. PMID: 20093390. PMCID: PMC2871726.9. Coll-Mulet L, Santidrián AF, Cosialls AM, et al. 2008; Multiplex ligation-dependent probe amplification for detection of genomic alterations in chronic lymphocytic leukaemia. Br J Haematol. 142:793–801. DOI: 10.1111/j.1365-2141.2008.07268.x. PMID: 18564355.
Article10. Buijs A, Krijtenburg PJ, Meijer E. 2006; Detection of risk-identifying chromosomal abnor-malities and genomic profiling by multiplex ligation-dependent probe amplification in chronic lymphocytic leukemia. Haematologica. 91:1434–5. PMID: 17018399.11. Alhourani E, Rincic M, Othman MA, et al. 2014; Comprehensive chronic lymphocytic leu-kemia diagnostics by combined multiplex ligation dependent probe amplification (MLPA) and interphase fluorescence in situ hybridization (iFISH). Mol Cytogenet. 7:79. DOI: 10.1186/s13039-014-0079-2. PMID: 25435911. PMCID: PMC4247644.
Article12. Hallek M, Cheson BD, Catovsky D, et al. 2018; iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 131:2745–60. DOI: 10.1182/blood-2017-09-806398. PMID: 29540348.
Article13. Sindelárová L, Michalová K, Zemanová Z, et al. 2005; Incidence of chromosomal anomalies detected with FISH and their clinical correlations in B-chronic lymphocytic leukemia. Cancer Genet Cytogenet. 160:27–34. DOI: 10.1016/j.cancergencyto.2004.11.004. PMID: 15949567.14. Patkar N, Rabade N, Kadam PA, et al. 2017; Immunogenetics of chronic lymphocytic leu-kemia. Indian J Pathol Microbiol. 60:38–42. DOI: 10.4103/IJPM.IJPM_466_16. PMID: 28195089.15. Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. 2008; Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 68:1012–21. DOI: 10.1158/0008-5472.CAN-07-3105. PMID: 18281475.
Article16. Ouillette P, Fossum S, Parkin B, et al. 2010; Aggressive chronic lymphocytic leukemia with elevated genomic complexity is associated with multiple gene defects in the response to DNA double-strand breaks. Clin Cancer Res. 16:835–47. DOI: 10.1158/1078-0432.CCR-09-2534. PMID: 20086003. PMCID: PMC2818663.
Article17. Ouillette P, Collins R, Shakhan S, et al. 2011; Acquired genomic copy number aberrations and survival in chronic lymphocytic leukemia. Blood. 118:3051–61. DOI: 10.1182/blood-2010-12-327858. PMID: 21795749. PMCID: PMC3175782.
Article18. Garg R, Wierda W, Ferrajoli A, et al. 2012; The prognostic difference of monoallelic versus biallelic deletion of 13q in chronic lymphocytic leukemia. Cancer. 118:3531–7. DOI: 10.1002/cncr.26593. PMID: 22139735. PMCID: PMC4535912.
Article19. Balatti V, Bottoni A, Palamarchuk A, et al. 2012; NOTCH1 mutations in CLL associated with trisomy 12. Blood. 119:329–31. DOI: 10.1182/blood-2011-10-386144. PMID: 22086416. PMCID: PMC3257004.
Article20. Abdool A, Donahue AC, Wohlgemuth JG, Yeh CH. 2010; Detection, analysis and clinical validation of chromosomal aberrations by multiplex ligation-dependent probe ampli-fication in chronic leukemia. PLoS One. 5:e15407. DOI: 10.1371/journal.pone.0015407. PMID: 21049055. PMCID: PMC2963645.
Article21. Cramer P, Hallek M. 2011; Prognostic factors in chronic lymphocytic leukemia-what do we need to know? Nat Rev Clin Oncol. 8:38–47. DOI: 10.1038/nrclinonc.2010.167. PMID: 20956983.
Article22. Delgado J, Espinet B, Oliveira AC, et al. 2012; Chronic lymphocytic leukaemia with 17p deletion: a retrospective analysis of prognostic factors and therapy results. Br J Haematol. 157:67–74. DOI: 10.1111/j.1365-2141.2011.09000.x. PMID: 22224845.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Recent advances in chronic lymphocytic leukemia therapy
- First Case of Transformation of Immunoglobulin Heavy Chain Variable-Mutated Chronic Lymphocytic Leukemia Into Chronic Myeloid Leukemia
- A Case of Leukemia Cutis Associated with B-cell Chronic Lymphocytic Leukemia
- Leukemic Macrocheilitis Associated with Chronic Lymphocytic Leukemia
- A case of B cell chronic lymphocytic leukemia involving thyroidgland