Ann Clin Microbiol.
2020 Mar;23(2):73-80. 10.5145/ACM.2020.23.2.4.
Killing Dynamics of Carbapenems against Pseudomonas aeruginosa Harboring Varied Determinants of Carbapenem Resistance
- Affiliations
-
- 1Global Health Security, Graduate School of Public Health, Yonsei University, Seoul, Korea
- 2Sangji University College of Science, Wonju, Korea
- 3Department of Laboratory Medicine and Research Institute of Bacterial Resistance, Yonsei University, College of Medicine, Seoul, Korea
- KMID: 2506546
- DOI: http://doi.org/10.5145/ACM.2020.23.2.4
Abstract
- Background
Ideal dose of the antimicrobials should be decided by considering their killing dynamics since sufficient elimination of the causative microorganisms is critical for proper antimicrobial treatment. In this study, the bactericidal activities of carbapenems by resistance mechanisms were assessed for carbapenem-resistant Pseudomonas aeruginosa .
Methods
Minimal inhibition concentrations (MICs) of carbapenems were determined by broth dilution method and the resistance mechanisms were identified by PCR and DNA sequencing. The expression levels of efflux pumps were determined by reverse transcriptase real-time PCR. Time-kill curves were plotted by time-course numeration of the viable cells grown under imipenem and meropenem at 1× and 4×MICs, respectively.
Results
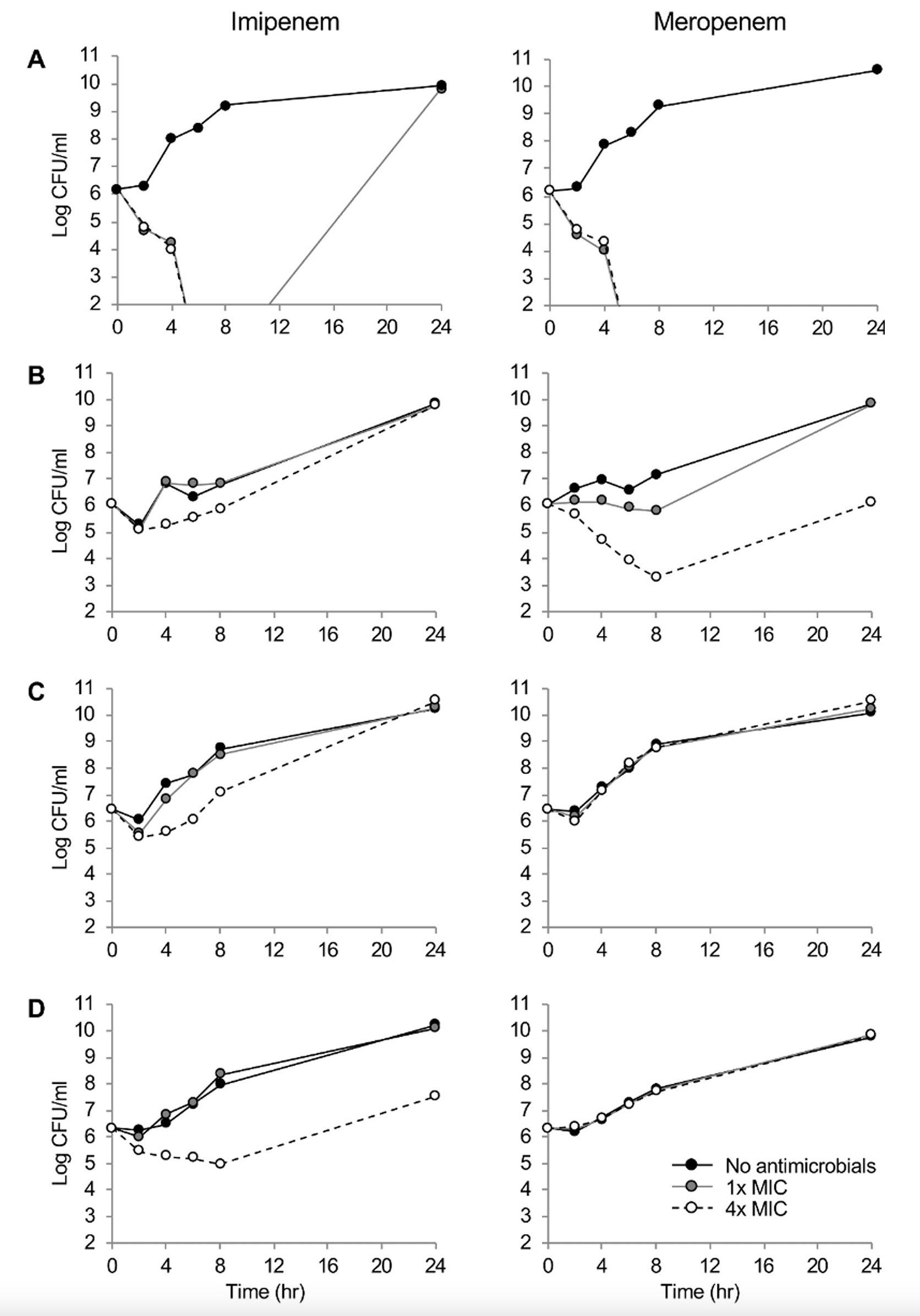
One P. aeruginosa strain was susceptible, whereas three were resistant to carbapenems by defective OprD, efflux pump overproduction, and/or IMP-6 production. The susceptible strain had imipenem and meropenem MICs of 2 and 1 mg/L, respectively. The MICs were elevated by eight-fold by defective OprD, 16- and 32-fold by the pump overproduction, and four- and >64-fold by the combination of two determinants and the IMP-6 carbapenemase. While both the carbapenems showed time-dependent bactericidal activity to the susceptible isolate, either of the carbapenem-resistant determinants, such as decreased membrane permeability, carbapenemase production, or the defective OprD, presented concentration-dependent bacteriostatic activity.
Conclusion
Different killing dynamics of the carbapenems were observed depending on the resistance determinants, and the results would guide a proper treatment strategy for the patients using these drugs.
Figure
Reference
-
1. Poole K. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2011;2:65.2. Lee H, Yoon EJ, Kim D, Jeong SH, Won EJ, Shin JH, et al. Antimicrobial resistance of major clinical pathogens in South Korea, May 2016 to April 2017: first one-year report from KorGLASS. Euro Surveill 2018;23:1800047.3. Livermore DM. Of Pseudomonas, porins, pumps and carbapenems. J Antimicrob Chemother 2001;47:247-50.4. Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Epidemiology and characteristics of metallo-beta-lactamase-producing Pseudomonas aeruginosa. Infect Chemother 2015;47:81-97. .5. Quale J, Bratu S, Gupta J, Landman D. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 2006;50:1633-41.6. Mattoes HM, Kuti JL, Drusano GL, Nicolau DP. Optimizing antimicrobial pharmacodynamics: dosage strategies for meropenem. Clin Ther 2004;26:1187-98.7. Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. CLSI Document M100S. 26th ed. Wayne; PA: 2016.8. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 2011;17:1791-8.9. Hong JS, Yoon EJ, Lee H, Jeong SH, Lee K. Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother 2016;60:7216-23.10. Stratton CW. Dead bugs don't mutate: susceptibility issues in the emergence of bacterial resistance. Emerg Infect Dis 2003;9:10-6.11. Pankey GA and Sabath LD. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of gram-positive bacterial infections. Clin Infect Dis 2004;38:864-70.12. Dantas RC, Ferreira ML, Gontijo-Filho PP, Ribas RM. Pseudomonas aeruginosa bacteraemia: independent risk factors for mortality and impact of resistance on outcome. J Med Microbiol 2014;63:1679-87.13. Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009-11 in14 European and Mediterranean countries. J Antimicrob Chemother 2014;69:1804-14.14. Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. Dissemination of IMP-6 metallo-betalactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J Antimicrob Chemother 2011;66:2791-6.15. Huang H and Hancock RE. Genetic definition of the substrate selectivity of outer membrane porin protein OprD of Pseudomonas aeruginosa. J Bacteriol 1993;175:7793-800.16. Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2000;44:3322-7.17. Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. Plasmid-encoded metallobeta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother 2001;45:1343-8.18. Nordmann P, Naas T, Fortineau N, Poirel L. Superbugs in the coming new decade; multidrug resistance and prospects for treatment of Staphylococcus aureus, Enterococcus spp. and Pseudomonas aeruginosa in 2010. Curr Opin Microbiol 2007;10:436-40.19. Woodford N, Turton JF, Livermore DM. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 2011;35:736-55.20. Yoon EJ, Kim D, Lee H, Lee HS, Shin JH, Park YS, et al. Mortality dynamics of Pseudomonas aeruginosa bloodstream infections and the influence of defective OprD on mortality: prospective observational study. J Antimicrob Chemother 2019;74:2774-83.21. Alcalde-Rico M, Hernando-Amado S, Blanco P, Martinez JL. Multidrug efflux pumps at the crossroad between antibiotic resistance and bacterial virulence. Front Microbiol 2016;7:1483.22. Yoon EJ, Balloy V, Fiette L, Chignard M, Courvalin P, Grillot-Courvalin C. Contribution of the Ade resistance-nodulation-cell division-type efflux pumps to fitness and pathogenesis of Acinetobacter baumannii. mBio 2016;7:e00697-16.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Antimicrobial Resistance and Clones of Acinetobacter Species and Pseudomonas aeruginosa
- Dissemination of Carbapenem-Resistance among Multidrug Resistant Pseudomonas aeruginosa carrying Metallo-Beta-Lactamase Genes, including the Novel bla(IMP-65) Gene in Thailand
- Epidemiology and Characteristics of Metallo-beta-Lactamase-Producing Pseudomonas aeruginosa
- Mechanisms of ceftolozane/tazobactam resistance in Pseudomonas aeruginosa isolates from South Korea: A diagrostic study
- Importance of the Molecular Epidemiological Monitoring of Carbapenem-Resistant Pseudomonas aeruginosa