Healthc Inform Res.
2018 Jul;24(3):170-178. 10.4258/hir.2018.24.3.170.
Fast Convolutional Method for Automatic Sleep Stage Classification
- Affiliations
-
- 1Machine Learning and Computer Vision (MLCV) Lab, Faculty of Computer Science, Universitas Indonesia, Jawa Barat, Indonesia. intanurma@gmail.com
- 2Department of Computer Science, Universitas Padjadjaran, Sumedang, Indonesia.
- KMID: 2418167
- DOI: http://doi.org/10.4258/hir.2018.24.3.170
Abstract
OBJECTIVES
Polysomnography is essential to diagnose sleep disorders. It is used to identify a patient's sleep pattern during sleep. This pattern is obtained by a doctor or health practitioner by using a scoring process, which is time consuming. To overcome this problem, we developed a system that can automatically classify sleep stages.
METHODS
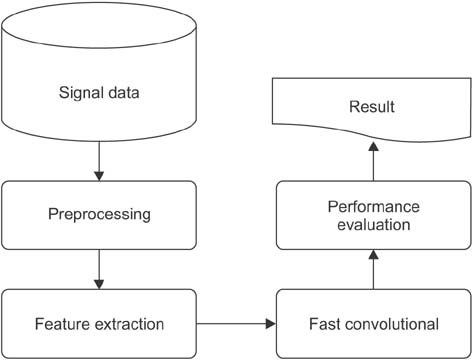
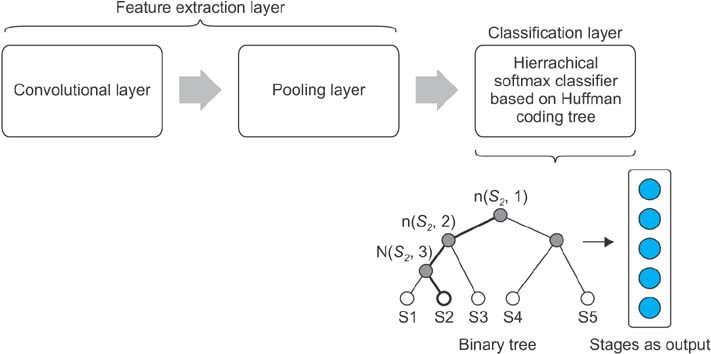
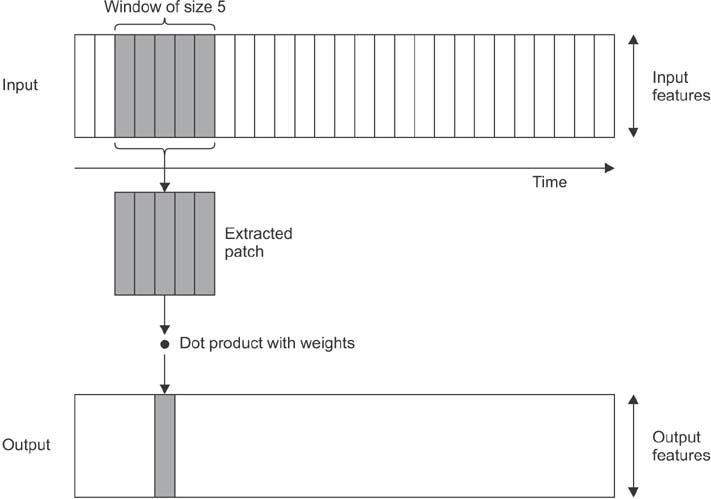
This paper proposes a new method for sleep stage classification, called the fast convolutional method. The proposed method was evaluated against two sleep datasets. The first dataset was obtained from physionet.org, a physiologic signals data centers. Twenty-five patients who had a sleep disorder participated in this data collection. The second dataset was collected in Mitra Keluarga Kemayoran Hospital, Indonesia. Data was recorded from ten healthy respondents.
RESULTS
The proposed method reached 73.50% and 56.32% of the F-measures for the PhysioNet and Mitra Keluarga Kemayoran Hospital data, respectively. Both values were the highest among all the machine learning methods considered in this study. The proposed method also had an efficient running time. The fast convolutional models of the PhysioNet and Mitra Keluarga Kemayoran Hospital data needed 42.60 and 0.06 seconds, respectively.
CONCLUSIONS
The fast convolutional method worked well on the tested datasets. It achieved a high F-measure result and an efficient running time. Thus, it can be considered a promising tool for sleep stage classification.
MeSH Terms
Figure
Reference
-
1. Hartzler BM. Fatigue on the flight deck: the consequences of sleep loss and the benefits of napping. Accid Anal Prev. 2014; 62:309–318.
Article2. McEwen BS, Karatsoreos IN. Sleep deprivation and circadian disruption: stress, allostasis, and allostatic load. Sleep Med Clin. 2015; 10(1):1–10.
Article3. Fullagar HH, Skorski S, Duffield R, Hammes D, Coutts AJ, Meyer T. Sleep and athletic performance: the effects of sleep loss on exercise performance, and physiological and cognitive responses to exercise. Sports Med. 2015; 45(2):161–186.
Article4. Weber F, Hoang Do JP, Chung S, Beier KT, Bikov M, Saffari Doost M, et al. Regulation of REM and Non-REM sleep by periaqueductal GABAergic neurons. Nat Commun. 2018; 9(1):354.
Article5. Begic D. “Polysomnographic” and “sleep” patterns: synonims or two distinct terms. Psychiatr Danub. 2015; 27(1):73.6. Kaditis A, Kheirandish-Gozal L, Gozal D. Pediatric OSAS: oximetry can provide answers when polysomnography is not available. Sleep Med Rev. 2016; 27:96–105.
Article7. de Raaff CA, Pierik AS, Coblijn UK, de Vries N, Bonjer HJ, van Wagensveld BA. Value of routine polysomnography in bariatric surgery. Surg Endosc. 2017; 31(1):245–248.
Article8. Boostani R, Karimzadeh F, Nami M. A comparative review on sleep stage classification methods in patients and healthy individuals. Comput Methods Programs Biomed. 2017; 140:77–91.
Article9. Aboalayon KA, Faezipour M, Almuhammadi WS, Moslehpour S. Sleep stage classification using EEG signal analysis: a comprehensive survey and new investigation. Entropy. 2016; 18(9):272.
Article10. Hassan AR, Bhuiyan MI. Computer-aided sleep staging using complete ensemble empirical mode decomposition with adaptive noise and bootstrap aggregating. Biomed Signal Process Control. 2016; 24:1–10.
Article11. Hassan AR, Subasi A. A decision support system for automated identification of sleep stages from singlechannel EEG signals. Knowl Based Syst. 2017; 128:115–124.
Article12. Kuo CY, Yu LC, Chen HC, Chan CL. Comparison of models for the prediction of medical costs of spinal fusion in Taiwan Diagnosis-Related Groups by machine learning algorithms. Healthc Inform Res. 2018; 24(1):29–37.
Article13. Langkvist M, Karlsson L, Loutfi A. A review of unsupervised feature learning and deep learning for time-series modeling. Pattern Recognit Lett. 2014; 42:11–24.
Article14. Gaur P, Pachori RB, Wang H, Prasad G. A multi-class EEG-based BCI classification using multivariate empirical mode decomposition based filtering and Riemannian geometry. Expert Syst Appl. 2018; 95:201–211.
Article15. Han D, An S, Shi P. Multi-frequency weak signal detection based on wavelet transform and parameter compensation band-pass multi-stable stochastic resonance. Mech Syst Signal Process. 2016; 70:995–1010.
Article16. Yener SC, Uygur A, Kuntman HH. Ultra low-voltage ultra low-power memristor based band-pass filter design and its application to EEG signal processing. Analog Integr Circuits Signal Process. 2016; 89(3):719–726.
Article17. Das A. Fast Fourier transform. In : Das A, editor. Signal conditioning: an introduction to continuous wave communication and signal processing. Heidelberg: Springer;2012. p. 193–215.18. Joulin A, Grave E, Bojanowski P, Douze M, Jegou H, Mikolov T. FastText.zip: compressing text classification models [Internet]. Ithaca (NY): arXiv.org;2016. cited at 2018 Jul 15. Available from: https://arxiv.org/abs/1612.03651.19. Chollet F. Deep learning with Python. Shelter Island (NY): Manning Publications Co.;2018.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Automatic disease stage classification of glioblastoma multiforme histopathological images using deep convolutional neural network
- Automatic Detection of Stage 1 Sleep Utilizing Simultaneous Analyses of EEG Spectrum and Slow Eye Movement
- Obstructive sleep apnoea detection using convolutional neural network based deep learning framework
- Automatic Sleep stage Scoring Using Hybrid Neural Network and Rule-based Expert Reasoning
- Respiratory Sleep Physiology