Korean J Lab Med.
2008 Dec;28(6):401-412. 10.3343/kjlm.2008.28.6.401.
Extended-spectrum beta-Lactamases: Implications for the Clinical Laboratory and Therapy
- Affiliations
-
- 1Department of Microbiology and Infectious Diseases, School of Medicine, Faculty of Medicine, Toho University, Tokyo, Japan. yishii@med.toho-u.ac.jp
- 2Division of Infectious Diseases, The University of Tokyo Hospital, Tokyo, Japan.
- KMID: 1781573
- DOI: http://doi.org/10.3343/kjlm.2008.28.6.401
Abstract
- Production of extended-spectrum beta-lactamase (ESBL) is one of the most important resistance mechanisms that hamper the antimicrobial treatment of infections caused by Enterobacteriaceae. ESBLs are classified into several groups according to their amino-acid sequence homology. While TEM and SHV enzymes were the most common ESBLs in the 1990s, CTX-M enzymes have spread rapidly among Enterobacteriaceae in the past decade. In addition, some epidemiological studies showed that organisms producing CTX-M enzymes had become increasingly prevalent in the community setting in certain areas in the world. Several novel enzymes with hydrolyzing activity against oxyimino-cephalosporins, albeit with additional enzymatic characteristics different from those of original TEM and SHV ESBLs (e.g., inhibitor-resistance), have been discovered and pose a problem on the definition of ESBLs. Although several methods to detect the production of ESBL are available in clinical laboratories, existence of other factors contributing resistance against beta-lactams, e.g., inducible production of Amp-C beta-lactamase by some species of Enterobacteriaceae, or inhibitor-resistance in some ESBLs may hinder the detection of ESBLs with these methods. Carbapenems are stable against hydrolyzing activity of ESBLs and are regarded as the drug of choice for the treatment of infections caused by ESBL-producing Enterobacteriaceae. Although several other antimicrobial agents, such as fluoroquinolones and cephamycins, may have some role in the treatment of mild infections due to those organisms, clinical data that warrant the use of antimicrobial agents other than carbapenems in the treatment of serious infections due to those organisms are scarce for now.
Keyword
MeSH Terms
-
Anti-Bacterial Agents/*pharmacology/therapeutic use
Carbapenems/pharmacology/therapeutic use
Disk Diffusion Antimicrobial Tests
Enterobacteriaceae/drug effects/*enzymology/genetics
Enterobacteriaceae Infections/*drug therapy/microbiology
Fluoroquinolones/pharmacology/therapeutic use
Humans
Microbial Sensitivity Tests/methods
beta-Lactamases/*biosynthesis/metabolism
beta-Lactams/*pharmacology/therapeutic use
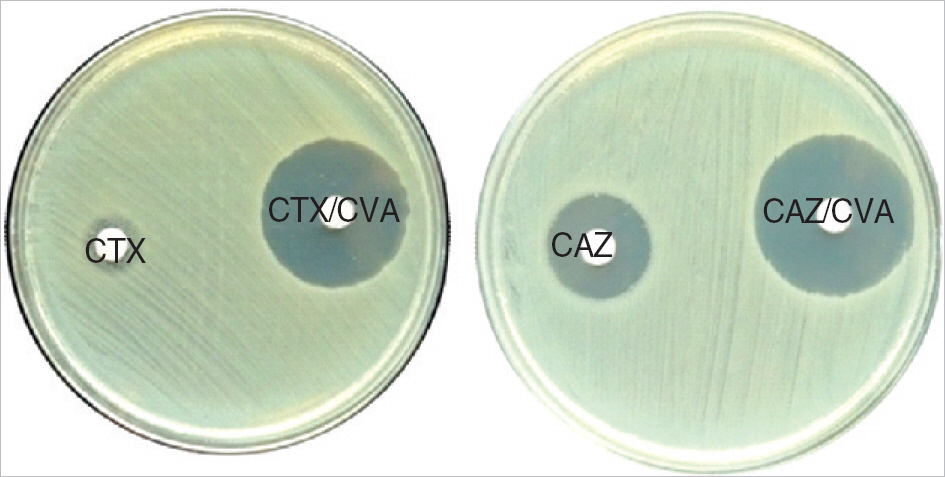
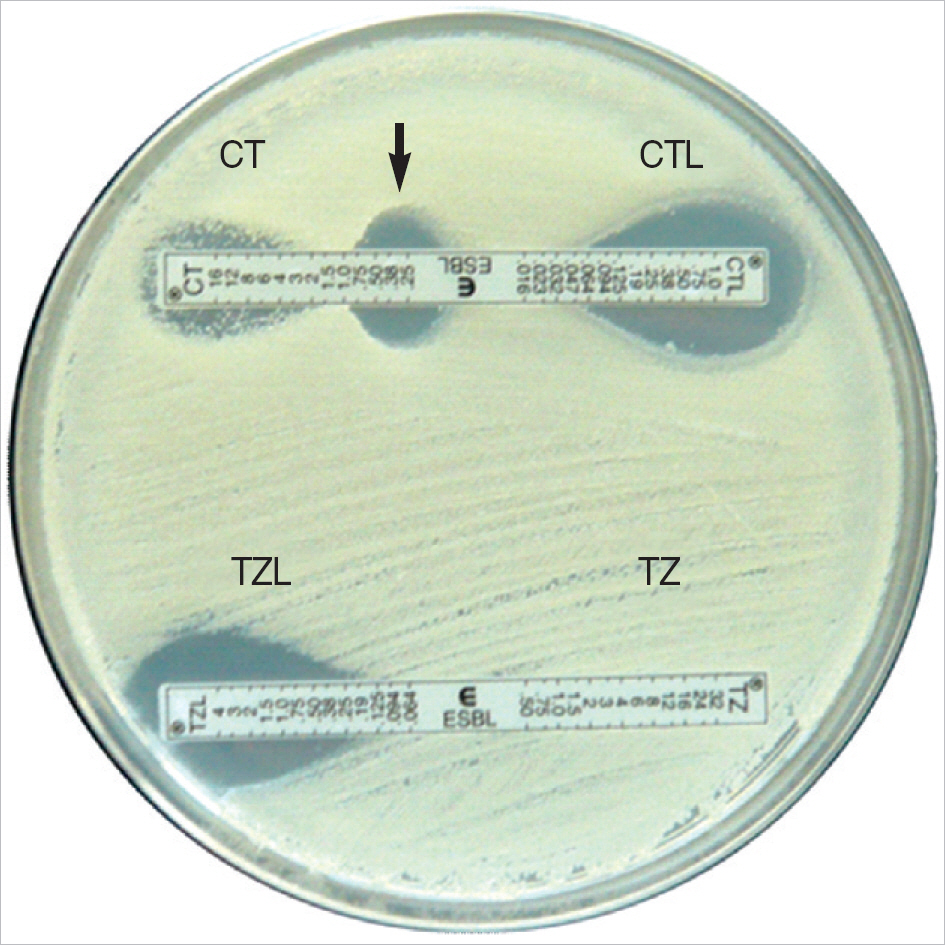
Figure
Reference
-
1.Livermore DM. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995. 8:557–84.2.Datta N., Kontomichalou P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature. 1965. 208:239–41.3.Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983. 11:315–7.4.Jacoby G., Bush K. Lahey clinic page on amino acid sequence for TEM, SHV and OXA extended-spectrum and inhibitor resistant β-Lactamases. http://www.lahey.org/Studies/. (Updated on Sep 24,. 2008.5.Bernard H., Tancrede C., Livrelli V., Morand A., Barthelemy M., Labia R. A novel plasmid-mediated extended-spectrum β-lactamase not derived from TEM- or SHV-type enzymes. J Antimicrob Chemother. 1992. 29:590–2.6.Canton R., Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006. 9:466–75.7.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004. 48:1–14.
Article8.Rossolini GM., D'Andrea MM., Mugnaioli C. The spread of CTX-M-type extended-spectrum β-lactamases. Clin Microbiol Infect. 2008. 14(S):S33–41.9.Ambler RP. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980. 289:321–31.10.Bush K., Jacoby GA., Medeiros AA. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995. 39:1211–33.11.Livermore DM. Defining an extended-spectrum β-lactamase. Clin Microbiol Infect. 2008. 14(S):S3–10.12.Livermore DM. Clinical significance of β-lactamase induction and stable derepression in gram-negative rods. Eur J Clin Microbiol. 1987. 6:439–45.13.Philippon A., Arlet G., Jacoby GA. Plasmid-determined AmpC-type β-lactamases. Antimicrob Agents Chemother. 2002. 46:1–11.
Article14.Mammeri H., Poirel L., Bemer P., Drugeon H., Nordmann P. Resistance to cefepime and cefpirome due to a 4-amino-acid deletion in the chromosome-encoded AmpC β-lactamase of a Serratia marcescens clinical isolate. Antimicrob Agents Chemother. 2004. 48:716–20.15.Barnaud G., Benzerara Y., Gravisse J., Raskine L., Sanson-Le Pors MJ., Labia R, et al. Selection during cefepime treatment of a new cephalosporinase variant with extended-spectrum resistance to cefepime in an Enterobacter aerogenes clinical isolate. Antimicrob Agents Chemother. 2004. 48:1040–2.16.Mammeri H., Nazic H., Naas T., Poirel L., Leotard S., Nordmann P. AmpC β-lactamase in an Escherichia coli clinical isolate confers resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 2004. 48:4050–3.17.Wachino J., Kurokawa H., Suzuki S., Yamane K., Shibata N., Kimura K, et al. Horizontal transfer of blaCMY-bearing plasmids among clinical Escherichia coli and Klebsiella pneumoniae isolates and emergence of cefepime-hydrolyzing CMY-19. Antimicrob Agents Chemother. 2006. 50:534–41.18.Kim JY., Jung HI., An YJ., Lee JH., Kim SJ., Jeong SH, et al. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C β-lactamase. Mol Microbiol. 2006. 60:907–16.19.Queenan AM., Bush K. Carbapenemases: the versatile β-lacta-mases. Clin Microbiol Rev. 2007. 20:440–58.
Article20.Poirel L., Le Thomas I., Naas T., Karim A., Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000. 44:622–32.21.Poirel L., Weldhagen GF., Naas T., De Champs C., Dove MG., Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001. 45:2598–603.22.Wachino J., Doi Y., Yamane K., Shibata N., Yagi T., Kubota T, et al. Molecular characterization of a cephamycin-hydrolyzing and inhibitor-resistant class A β-lactamase, GES-4, possessing a single G170S substitution in the omega-loop. Antimicrob Agents Chemother. 2004. 48:2905–10.23.Matthew M., Hedges RW., Smith JT. Types of β-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979. 138:657–62.24.Babini GS., Livermore DM. Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob Agents Chemother. 2000. 44:2230.25.Kliebe C., Nies BA., Meyer JF., Tolxdorff-Neutzling RM., Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985. 28:302–7.
Article26.Heritage J., M'Zali FH., Gascoyne-Binzi D., Hawkey PM. Evolution and spread of SHV extended-spectrum β-lactamases in gram-negative bacteria. J Antimicrob Chemother. 1999. 44:309–18.
Article27.Huletsky A., Knox JR., Levesque RC. Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type β-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J Biol Chem. 1993. 268:3690–7.28.Ford PJ., Avison MB. Evolutionary mapping of the SHV β-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother. 2004. 54:69–75.29.Miriagou V., Carattoli A., Tzelepi E., Villa L., Tzouvelekis LS. IS26-associated In4-type integrons forming multiresistance loci in enter-obacterial plasmids. Antimicrob Agents Chemother. 2005. 49:3541–3.30.Preston KE., Venezia RA., Stellrecht KA. The SHV-5 extended-spectrum β-lactamase gene of pACM1 is located on the remnant of a compound transposon. Plasmid. 2004. 51:48–53.
Article31.Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001. 14:933–51.
Article32.Sirot D., Sirot J., Labia R., Morand A., Courvalin P., Darfeuille-Michaud A, et al. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel β-lactamase. J Antimicrob Chemother. 1987. 20:323–34.33.Sougakoff W., Goussard S., Courvalin P. The TEM-3 β-lactamase, which hydrolyzes broad-spectrum cephalosporins, is derived from the TEM-2 penicillinase by two amino acid substitutions. FEMS Microbiol Lett. 1988. 56:343–8.
Article34.Canton R., Morosini MI., de la Maza OM., de la Pedrosa EG. IRT and CMT β-lactamases and inhibitor resistance. Clin Microbiol Infect. 2008. 14(S):S53–62.35.Robin F., Delmas J., Archambaud M., Schweitzer C., Chanal C., Bonnet R. CMT-type β-lactamase TEM-125, an emerging problem for extended-spectrum β-lactamase detection. Antimicrob Agents Chemother. 2006. 50:2403–8.36.Bauernfeind A., Grimm H., Schweighart S. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection. 1990. 18:294–8.37.Barthelemy M., Peduzzi J., Bernard H., Tancrede C., Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992. 1122:15–22.38.Bauernfeind A., Casellas JM., Goldberg M., Holley M., Jungwirth R., Mangold P, et al. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992. 20:158–63.39.Ishii Y., Ohno A., Taguchi H., Imajo S., Ishiguro M., Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995. 39:2269–75.40.Bauernfeind A., Stemplinger I., Jungwirth R., Ernst S., Casellas JM. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996. 40:509–13.41.Humeniuk C., Arlet G., Gautier V., Grimont P., Labia R., Philippon A. β-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob Agents Chemother. 2002. 46:3045–9.42.Poirel L., Kampfer P., Nordmann P. Chromosome-encoded Ambler class A β-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2002. 46:4038–40.43.Rodriguez MM., Power P., Radice M., Vay C., Famiglietti A., Galleni M, et al. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob Agents Chemother. 2004. 48:4895–7.44.Olson AB., Silverman M., Boyd DA., McGeer A., Willey BM., Pong-Porter V, et al. Identification of a progenitor of the CTX-M-9 group of extended-spectrum β-lactamases from Kluyvera georgiana isolated in Guyana. Antimicrob Agents Chemother. 2005. 49:2112–5.45.Lartigue MF., Poirel L., Aubert D., Nordmann P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring β-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob Agents Chemother. 2006. 50:1282–6.46.Poirel L., Decousser JW., Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother. 2003. 47:2938–45.47.Rodriguez-Martinez JM., Poirel L., Canton R., Nordmann P. Common region CR1 for expression of antibiotic resistance genes. Antimicrob Agents Chemother. 2006. 50:2544–6.
Article48.Oliver A., Coque TM., Alonso D., Valverde A., Baquero F., Cantón R. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob Agents Chemother. 2005. 49:1567–71.49.Yu WL., Pfaller MA., Winokur PL., Jones RN. Cefepime MIC as a predictor of the extended-spectrum β-lactamase type in Klebsiella pneumoniae, Taiwan. Emerg Infect Dis. 2002. 8:522–4.50.Chen Y., Delmas J., Sirot J., Shoichet B., Bonnet R. Atomic resolution structures of CTX-M β-lactamases: extended spectrum activities from increased mobility and decreased stability. J Mol Biol. 2005. 348:349–62.
Article51.Ibuka AS., Ishii Y., Galleni M., Ishiguro M., Yamaguchi K., Frere JM, et al. Crystal structure of extended-spectrum β-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry. 2003. 42:10634–43.
Article52.Kimura S., Ishiguro M., Ishii Y., Alba J., Yamaguchi K. Role of a mutation at position 167 of CTX-M-19 in ceftazidime hydrolysis. Antimicrob Agents Chemother. 2004. 48:1454–60.
Article53.Ishii Y., Galleni M., Ma L., Frere JM., Yamaguchi K. Biochemical characterisation of the CTX-M-14 β-lactamase. Int J Antimicrob Agents. 2007. 29:159–64.54.Pitout JD., Nordmann P., Laupland KB., Poirel L. Emergence of Enter-obacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005. 56:52–9.55.Walther-Rasmussen J., Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006. 57:373–83.
Article56.Naas T., Poirel L., Nordmann P. Minor extended-spectrum β-lactamases. Clin Microbiol Infect. 2008. 14(S1):S42–52.
Article57.Paterson DL., Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005. 18:657–86.58.Paterson DL., Ko WC., Von Gottberg A., Casellas JM., Mulazimoglu L., Klugman KP, et al. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. J Clin Microbiol. 2001. 39:2206–12.
Article59.Siegel JD, Rhinehart E, Jackson M, Chiarello L, Health Infection Control Practices Advisory Committee. Management of multidrug-resistant organisms in healthcare settings. 2006. http://www.cdc.gov/ncidod/dhqp/pdf/ar/MDROGuideline2006.pdf.60.Kahlmeter G. Breakpoints for intravenously used cephalosporins in Enterobacteriaceae -EUCAST and CLSI breakpoints. Clin Microbiol Infect. 2008. 14(S):S169–74.61.Weber DA., Sanders CC. Diverse potential of β-lactamase inhibitors to induce class I enzymes. Antimicrob Agents Chemother. 1990. 34:156–8.62.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility tasting; 18th informational supplement. M100-S18. Wayne, PA: Clinical and Laboratory Standards Institute;2008.63.Brenwald NP., Jevons G., Andrews JM., Xiong JH., Hawkey PM., Wise R. An outbreak of a CTX-M-type β-lactamase-producing Klebsiella pneumoniae: the importance of using cefpodoxime to detect extended-spectrum β-lactamases. J Antimicrob Chemother. 2003. 51:195–6.64.Jarlier V., Nicolas MH., Fournier G., Philippon A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988. 10:867–78.65.Thomson KS., Sanders CC. Detection of extended-spectrum β-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother. 1992. 36:1877–82.66.Tzelepi E., Giakkoupi P., Sofianou D., Loukova V., Kemeroglou A., Tsakris A. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000. 38:542–6.67.Stürenburg E., Sobottka I., Noor D., Laufs R., Mack D. Evaluation of a new cefepime-clavulanate ESBL Etest to detect extended-spectrum β-lactamases in an Enterobacteriaceae strain collection. J Antimicrob Chemother. 2004. 54:134–8.68.Leverstein-van Hall MA., Fluit AC., Paauw A., Box AT., Brisse S., Verhoef J. Evaluation of the Etest ESBL and the BD Phoenix, VITEK 1, and VITEK 2 automated instruments for detection of extended-spectrum β-lactamases in multiresistant Escherichia coli and Klebsiella spp. J Clin Microbiol. 2002. 40:3703–11.69.Wiegand I., Geiss HK., Mack D., Stürenburg E., Seifert H. Detection of extended-spectrum β-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J Clin Microbiol. 2007. 45:1167–74.70.Thomson KS., Cornish NE., Hong SG., Hemrick K., Herdt C., Moland ES. Comparison of Phoenix and VITEK 2 extended-spectrum-β-lactamase detection tests for analysis of Escherichia coli and Klebsiella isolates with well-characterized beta-lactamases. J Clin Microbiol. 2007. 45:2380–4.71.Sanguinetti M., Posteraro B., Spanu T., Ciccaglione D., Romano L., Fiori B, et al. Characterization of clinical isolates of Enterobacteriaceae from Italy by the BD Phoenix extended-spectrum β-lactamase detection method. J Clin Microbiol. 2003. 41:1463–8.72.Spanu T., Sanguinetti M., Tumbarello M., D'Inzeo T., Fiori B., Posteraro B, et al. Evaluation of the new VITEK 2 extended-spectrum β-lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J Clin Microbiol. 2006. 44:3257–62.73.Stürenburg E., Sobottka I., Feucht HH., Mack D., Laufs R. Comparison of BD Phoenix and VITEK2 automated antimicrobial susceptibility test systems for extended-spectrum β-lactamase detection in Escherichia coli and Klebsiella species clinical isolates. Diagn Microbiol Infect Dis. 2003. 45:29–34.74.Jeong SH., Song W., Park MJ., Kim JS., Kim HS., Bae IK, et al. Boronic acid disk tests for identification of extended-spectrum β-lactamase production in clinical isolates of Enterobacteriaceae producing chromosomal AmpC β-lactamases. Int J Antimicrob Agents. 2008. 31:467–71.75.Song W., Bae IK., Lee YN., Lee CH., Lee SH., Jeong SH. Detection of extended-spectrum β-lactamases by using boronic acid as an AmpC β-lactamase inhibitor in clinical isolates of Klebsiella spp. and Escherichia coli. J Clin Microbiol. 2007. 45:1180–4.76.Pitout JD., Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008. 8:159–66.
Article77.Paterson DL., Ko WC., Von Gottberg A., Mohapatra S., Casellas JM., Goossens H, et al. Antibiotic therapy for Klebsiella pneumoniae bacteremia: implications of production of extended-spectrum β-lactamases. Clin Infect Dis. 2004. 39:31–7.78.Lautenbach E., Strom BL., Bilker WB., Patel JB., Edelstein PH., Fishman NO. Epidemiological investigation of fluoroquinolone resistance in infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Clin Infect Dis. 2001. 33:1288–94.79.Paterson DL., Mulazimoglu L., Casellas JM., Ko WC., Goossens H., Von Gottberg A, et al. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin Infect Dis. 2000. 30:473–8.80.Thomson KS., Moland ES. Cefepime, piperacillin-tazobactam, and the inoculum effect in tests with extended-spectrum β-lactamase-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2001. 45:3548–54.81.Bedenic B., Beader N., Zagar Z. Effect of inoculum size on the antibacterial activity of cefpirome and cefepime against Klebsiella pneumoniae strains producing SHV extended-spectrum β-lactamases. Clin Microbiol Infect. 2001. 7:626–35.82.Bin C., Hui W., Renyuan Z., Yongzhong N., Xiuli X., Yingchun X, et al. Outcome of cephalosporin treatment of bacteremia due to CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli. Diagn Microbiol Infect Dis. 2006. 56:351–7.83.Endimiani A., Luzzaro F., Perilli M., Lombardi G., Coli A., Tamborini A, et al. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum β-lactamase: treatment outcome of patients receiving imipenem or ciprofloxacin. Clin Infect Dis. 2004. 38:243–51.84.Kang CI., Kim SH., Park WB., Lee KD., Kim HB., Kim EC, et al. Bloodstream infections due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for mortality and treatment outcome, with special emphasis on antimicrobial therapy. Antimicrob Agents Chemother. 2004. 48:4574–81.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Management of Extended-Spectrum Beta-Lactamase-Positive Gram-Negative Bacterial Urologic Infections
- Two Cases of Neonatal Osteomyelitis due to Extended Spectrum beta-lactamase Producing Klebsiella pneumoniae
- A Case Report of Sepsis by Extended-Spectrum beta-Lactamase Producing Escherichia Coli
- Strategies for Interpretive Standards of beta-Lactams Susceptibility Testing and Identification of Extended-Spectrum beta-Lactamases and Carbapenemases in Enterobacteriaceae
- Detection Rate of Extended-Spectrum beta-Lactamase Producers in Klebsiella pneumoniae and Escherichia coli Isolated at Yeungnam University Medical Center