J Korean Med Sci.
2005 Jun;20(3):450-455. 10.3346/jkms.2005.20.3.450.
Identification of Proteins Differentially Expressed in the Conventional Renal Cell Carcinoma by Proteomic Analysis
- Affiliations
-
- 1Department of Urologym, College of Medicine Gyeongsang National University, Jinju, Korea. seogee@gaechuk.gsnu.ac.kr
- 2Department of Biochemistry, College of Medicine and Institute of Health Science, Gyeongsang National University, Jinju, Korea.
- KMID: 1778505
- DOI: http://doi.org/10.3346/jkms.2005.20.3.450
Abstract
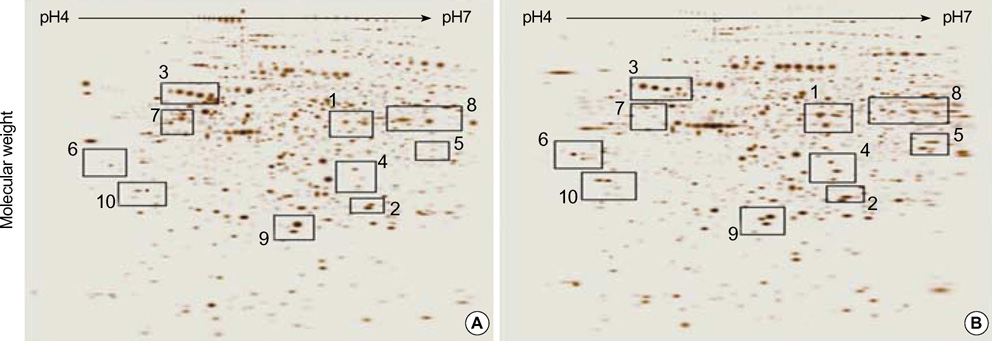
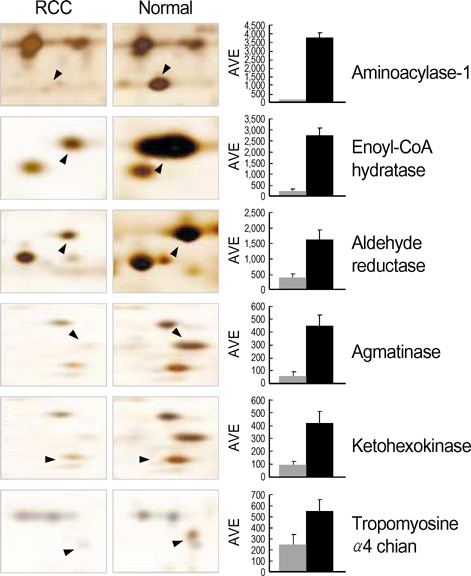
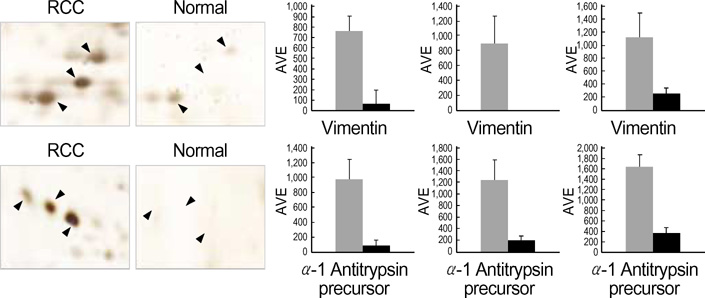
- Renal cell carcinoma (RCC) is one of the most malignant tumors in urology, and due to its insidious onset patients frequently have advanced disease at the time of clinical presentation. Thus, early detection is crucial in management of RCC. To identify tumor specific proteins of RCC, we employed proteomic analysis. We prepared proteins from conventional RCC and the corresponding normal kidney tissues from seven patients with conventional RCC. The expression of proteins was determined by silver stain after two-dimensional polyacrylamide gel electrophoresis (2D-PAGE). The overall protein expression patterns in the RCC and the normal kidney tissues were quite similar except some areas. Of 66 differentially expressed protein spots (p<0.05 by Student t-test), 8 different proteins from 11 spots were identified by MALDI-TOF-MS. The expression of the following proteins was repressed (p<0.05); aminoacylase-1, enoyl-CoA hydratase, aldehyde reductase, tropomyosin alpha-4 chain, agmatinase and ketohexokinase. Two proteins, vimentin and alpha-1 antitrypsin precursor, were dominantly expressed in RCC (p<0.05).
Keyword
MeSH Terms
-
Aged
Aldehyde Reductase/analysis
Amidohydrolases/analysis
Carcinoma, Renal Cell/*metabolism/pathology
Comparative Study
Electrophoresis, Gel, Two-Dimensional
Enoyl-CoA Hydratase/analysis
Female
Fructokinases/analysis
Humans
Kidney Neoplasms/*metabolism/pathology
Male
Middle Aged
Proteome/*analysis
Proteomics/*methods
Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization
Tropomyosin/analysis
Ureohydrolases/analysis
Vimentin/analysis
alpha 1-Antitrypsin/analysis
Figure
Cited by 1 articles
-
Disease-specific Proteins from Rheumatoid Arthritis Patients
Choong Won Kim, Eun Hye Cho, Yun Jong Lee, Yoon Hee Kim, Young-Sool Hah, Deok Ryong Kim
J Korean Med Sci. 2006;21(3):478-484. doi: 10.3346/jkms.2006.21.3.478.
Reference
-
1. Mydlo JH. Growth factors and renal cancer: characterization and therapeutic implications. World J Urol. 1995. 13:356–363.
Article2. Stenzl A, deKernion JB. The natural history of renal cell carcinoma. Semin Urol. 1989. 7:144–148.3. Pantuck AJ, Zisman A, Belldegrun AS. The changing natural history of renal cell carcinoma. J Urol. 2001. 166:1611–1623.
Article4. Vasavada SP, Novick AC, Williams BR. p53, bcl-2, and Bax expression in renal cell carcinoma. Urology. 1998. 51:1057–1061.
Article5. Ji H, Reid GE, Moritz RL, Eddes JS, Burgess AW, Simpson RJ. A two-dimensional gel database of human colon carcinoma proteins. Electrophoresis. 1997. 18:605–613.
Article6. Hirano T, Franzen B, Uryu K, Okuzawa K, Alaiya AA, Vanky F, Rodrigues L, Ebihara Y, Kato H, Auer G. Detection of polypeptides associated with the histopathological differentiation of primary lung carcinoma. Br J Cancer. 1995. 72:840–848.
Article7. Giometti CS, Williams K, Tollaksen SL. A two-dimensional electrophoresis database of human breast epithelial cell proteins. Electrophoresis. 1997. 18:573–581.
Article8. Sarto C, Marocchi A, Sanchez JC, Giannone D, Frutiger S, Golaz O, Wilkins MR, Doro G, Cappellano F, Hughes G, Hochstrasser DF, Mocarelli P. Renal cell carcinoma and normal kidney protein expression. Electrophoresis. 1997. 18:599–604.
Article9. Guinan P, Sobin LH, Algaba F, Badellino F, Kameyama S, MacLennan G, Novick A. TNM staging of renal cell carcinoma: Workgroup No. 3. Union International Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC). Cancer. 1997. 80:992–993.10. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976. 72:248–254.
Article11. Jin SH, Cho EH, Ko JE, Jung EH, Ahn B, Hahm JR, Kim JW, Kim CW, Kim DR. Comparative analysis of nuclear proteins of B cells in different developmental stages. Proteomics. 2003. 3:2428–2436.
Article12. Kellner R, Lichtenfels R, Atkins D, Bukur J, Ackermann A, Beck J, Brenner W, Melchior S, Seliger B. Targeting of tumor associated antigens in renal cell carcinoma using proteome-based analysis and their clinical significance. Proteomics. 2002. 2:1743–1751.
Article13. Balabanov S, Zimmermann U, Protzel C, Scharf C, Klebingat KJ, Walther R. Tumour-related enzyme alterations in the clear cell type of human renal cell carcinoma identified by two-dimensional gel electrophoresis. Eur J Biochem. 2001. 268:5977–5980.
Article14. Seliger B, Menig M, Lichtenfels R, Atkins D, Bukur J, Halder TM, Kersten M, Harder A, Ackermann A, Beck J, Muehlenweg B, Brenner W, Melchior S, Kellner R, Lottspeich F. Identification of markers for the selection of patients undergoing renal cellcarcinoma-specific immunotherapy. Proteomics. 2003. 3:979–990.15. Dallmann K, Junker H, Balabanov S, Zimmermann U, Giebel J, Walther R. Human agmatinase is diminished in the clear cell type of renal cell carcinoma. Int J Cancer. 2004. 108:342–347.
Article16. Droz D, Zachar D, Charbit L, Gogusev J, Chretein Y, Iris L. Expression of the human nephron differentiation molecules in renal cell carcinomas. Am J Pathol. 1990. 137:895–905.17. Lindner H, Hopfner S, Tafler-Naumann M, Miko M, Konrad L, Rohm KH. The distribution of aminoacylase I among mammalian species and localization of the enzyme in porcine kidney. Biochimie. 2000. 82:129–137.
Article18. Cook RM, Franklin WA, Moore MD, Johnson BE, Miller YE. Mutational inactivation of aminoacylase-I in a small cell lung cancer cell line. Genes Chromosomes Cancer. 1998. 21:320–325.19. Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, Schmidt L, Zhou F, Li H, Wei MH, Chen F, Glenn G, Choyke P, Walther MM, Weng Y, Duan DS, Dean M, Glavac D, Richards FM, Crossy PA, Ferguson-Smith MA, Le Paslier D, Chumakov I, Cohen D, Chinault AC, Maher ER, Linehan WM, Zbar B, Lerman MI. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993. 260:1317–1320.
Article20. Shuin T, Kondo K, Torigoe S, Kishida T, Kubota Y, Hosaka M, Nagashima Y, Kitamura H, Latif F, Zbar B, Lerman MI, Yao M. Frequent somatic mutations and loss of heterozygosity of the von Hippel-Lindau tumor suppressor gene in primary human renal cell carcinomas. Cancer Res. 1994. 54:2852–2855.21. Flynn TG. Aldehyde reductases: monomeric NADPH-dependent oxidoreductases with multifunctional potential. Biochem Pharmacol. 1982. 31:2705–2712.22. Barski OA, Gabbay KH, Grimshaw CE, Bohren KM. Mechanism of human aldehyde reductase: characterization of the active site pocket. Biochemistry. 1995. 34:11264–11275.
Article23. Cohen C, McCue PA, Derose PB. Histogenesis of renal cell carcinoma and renal oncocytoma. An immunohistochemical study. Cancer. 1988. 62:1946–1951.
Article24. Potempa J, Korzus E, Travis J. The serpin superfamily of proteinase inhibitors: structure, function, and regulation. J Biol Chem. 1994. 269:15957–15960.
Article25. Donaldson IA, Doyle TC, Matas N. Expression of rat liver ketohexokinase in yeast results in fructose intolerance. Biochem J. 1993. 291:179–186.
Article26. Tsuda H, Hacker HJ, Katayama H, Masui T, Ito N, Bannasch P. Correlative histochemical studies on preneoplastic and neoplastic lesions in the kidney of rats treated with nitrosamines. Virchows Arch B Cell Pathol Incl Mol Pathol. 1986. 51:385–404.
Article27. Singh M, Singh VN, August JT, Horecker BL. Alterations in glucose metabolism in chick embryo cells transformed by Rous sarcoma virus. Transformation-specific changes in the activities of key enzymes of the glycolytic and hexose monophosphate shunt pathways. Arch Biochem Biophys. 1974. 165:240–246.28. Beemer FA, Vlug AM, Rousseau-Merck MF, van Veelen CW, Rijksen G, Staal GE. Glycolytic enzymes from human neuroectodermal tumors of childhood. Eur J Cancer Clin Oncol. 1984. 20:253–259.
Article29. Beemer FA, Vlug AM, Rijksen G, Hamburg A, Staal GE. Characterization of some glycolytic enzymes from human retina and retinoblastoma. Cancer Res. 1982. 42:4228–4232.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The effect of 4-hexylresocinol administration on SCC-9 cells: mass spectrometric identification of proteins and cDNA microarray analysis
- Quantitative analysis of proteins related to chemoresistance to paclitaxel and carboplatin in human SiHa cervical cancer cells via iTRAQ
- Proteomic Analysis of Cellular and Membrane Proteins in Fluconazole-Resistant
Candida glabrata - Proteomic Comparison of Paraspinal Muscle Imbalance Between Idiopathic Scoliosis and Congenital Scoliosis
- Analysis of Gene Expression in Renal Cell Carcinomas Using cDNA Microarray: Reduced Expression of Decorin in Renal Cell Carcinomas