Brain Tumor Res Treat.
2014 Apr;2(1):22-28. 10.14791/btrt.2014.2.1.22.
Proteomic Analysis between U87MG and U343MG-A Cell Lines: Searching for Candidate Proteins for Glioma Invasion
- Affiliations
-
- 1Brain Tumor Research Laboratory, Department of Neurosurgery, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Hwasun, Korea. moonks@chonnam.ac.kr
- 2Department of Neurosurgery, Worker's Hospital of Tangshan, Tangshan City, China.
- 3Department of Chemistry, College of Life Science, Chonnam National University, Gwangju, Korea.
- 4Brain Tumor Research Laboratory, Department of Pathology, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Hwasun, Korea.
- KMID: 1734664
- DOI: http://doi.org/10.14791/btrt.2014.2.1.22
Abstract
- BACKGROUND
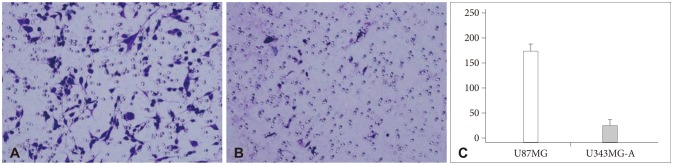
To investigate the molecular basis for invasion of malignant gliomas, proteomic analysis approach was carried out using two human glioma cell lines, U87MG and U343MG-A that demonstrate different motility and invasiveness in in vitro experiments.
METHODS
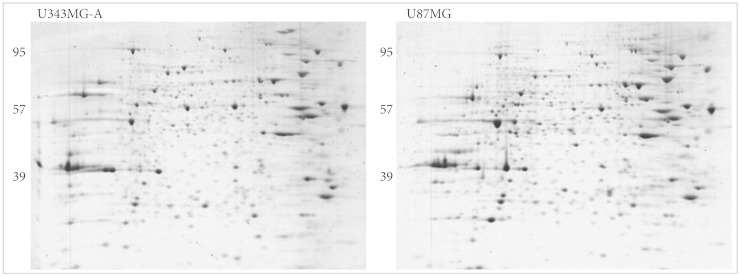
High-resolution two-dimensional gel electrophoresis and matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry analysis were performed.
RESULTS
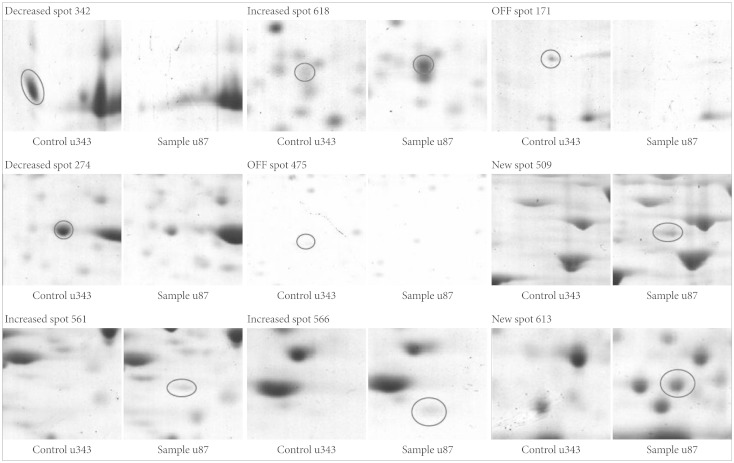
Nine distinct protein spots that were recognized with significant alteration between the two cell lines. Five of these protein spots were up-regulated in U87MG and four were up-regulated in U343MG-A.
CONCLUSION
Among these proteins, cathepsin D was shown to be one of the important proteins which are related with glioma invasion. However, further studies are necessary to reveal the exact role and mechanism of cathepsin D in glioma invasion.
Keyword
MeSH Terms
Figure
Reference
-
1. Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002; 415:436–442. PMID: 11807556.
Article2. Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001; 61:6885–6891. PMID: 11559565.3. Sallinen SL, Sallinen PK, Haapasalo HK, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000; 60:6617–6622. PMID: 11118044.4. Celis JE, Gromov P, Ostergaard M, et al. Human 2-D PAGE databases for proteome analysis in health and disease: http://biobase.dk/cgi-bin/celis. FEBS Lett. 1996; 398:129–134. PMID: 8977092.5. Godovac-Zimmermann J, Brown LR. Perspectives for mass spectrom-etry and functional proteomics. Mass Spectrom Rev. 2001; 20:1–57. PMID: 11223909.
Article6. Ryu HH, Jung S, Sun HS, et al. Screening for motility-associated genes in malignant astrocytoma cell lines. J Neurooncol. 2007; 82:125–131. PMID: 17048098.
Article7. Park SG, Jung S, Ryu HH, et al. Role of 14-3-3-beta in the migration and invasion in human malignant glioma cell line U87MG. Neurol Res. 2012; 34:893–900. PMID: 22925547.
Article8. Ryu HH, Jung S, Jung TY, et al. Role of metallothionein 1E in the migration and invasion of human glioma cell lines. Int J Oncol. 2012; 41:1305–1313. PMID: 22843066.
Article9. Jung TY, Jung S, Ryu HH, et al. Role of galectin-1 in migration and invasion of human glioblastoma multiforme cell lines. J Neurosurg. 2008; 109:273–284. PMID: 18671640.
Article10. Jung S, Paek YW, Moon KS, et al. Expression of Nm23 in gliomas and its effect on migration and invasion in vitro. Anticancer Res. 2006; 26:249–258. PMID: 16475705.11. Havlis J, Thomas H, Sebela M, Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem. 2003; 75:1300–1306. PMID: 12659189.12. Maruo T, Ichikawa T, Kanzaki H, et al. Proteomics-based analysis of invasion-related proteins in malignant gliomas. Neuropathology. 2013; 33:264–275. PMID: 23116197.
Article13. Diment S, Martin KJ, Stahl PD. Cleavage of parathyroid hormone in macrophage endosomes illustrates a novel pathway for intracellular processing of proteins. J Biol Chem. 1989; 264:13403–13406. PMID: 2760027.
Article14. Yin L, Stearns R, González-Flecha B. Lysosomal and mitochondrial pathways in H2O2-induced apoptosis of alveolar type II cells. J Cell Biochem. 2005; 94:433–445. PMID: 15534871.
Article15. Sagulenko V, Muth D, Sagulenko E, Paffhausen T, Schwab M, Westermann F. Cathepsin D protects human neuroblastoma cells from doxorubicin-induced cell death. Carcinogenesis. 2008; 29:1869–1877. PMID: 18566016.
Article16. Berchem G, Glondu M, Gleizes M, et al. Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene. 2002; 21:5951–5955. PMID: 12185597.
Article17. Hah YS, Noh HS, Ha JH, et al. Cathepsin D inhibits oxidative stress-induced cell death via activation of autophagy in cancer cells. Cancer Lett. 2012; 323:208–214. PMID: 22542809.
Article18. Fukuda ME, Iwadate Y, Machida T, et al. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005; 65:5190–5194. PMID: 15958563.
Article19. Iwadate Y, Sakaida T, Hiwasa T, et al. Molecular classification and survival prediction in human gliomas based on proteome analysis. Cancer Res. 2004; 64:2496–2501. PMID: 15059904.
Article20. Sun S, Wong TS, Zhang XQ, et al. Protein alterations associated with temozolomide resistance in subclones of human glioblastoma cell lines. J Neurooncol. 2012; 107:89–100. PMID: 21979894.
Article21. Liu Y, Zhou Y, Zhu K. Inhibition of glioma cell lysosome exocytosis inhibits glioma invasion. PLoS One. 2012; 7:e45910. PMID: 23029308.
Article22. Wu X, Hu A, Zhang M, Chen Z. Effects of Rab27a on proliferation, invasion, and anti-apoptosis in human glioma cell. Tumour Biol. 2013; 34:2195–2203. PMID: 23553027.
Article23. Dyer LM, Schooler KP, Ai L, et al. The transglutaminase 2 gene is aberrantly hypermethylated in glioma. J Neurooncol. 2011; 101:429–440. PMID: 20596752.
Article24. Yao X, Li D, Xiong DM, Li L, Jiang R, Chen JX. A novel role of ribonuclease inhibitor in regulation of epithelial-to-mesenchymal transition and ILK signaling pathway in bladder cancer cells. Cell Tissue Res. 2013; 353:409–423. PMID: 23703635.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Effect of Hyaluronic Acid on the Invasiveness of Malignant Glioma Cells : Comparison of Invasion Potential at Hyaluronic Acid Hydrogel and Matrigel
- Effects of Hydroxychloroquine Co-administered with Chemotherapeutic Agents on Malignant Glioma Cell Lines: in vitro Study
- The PPARgamma Agonist Rosiglitazone Inhibits Glioma Cell Proliferation and Migration in vitro and Glioma Tumor Growth in vivo
- Inhibitory Effects of Toxoplasma Antigen on Proliferation and Invasion of Human Glioma Cells
- High Expression of KIFC1 in Glioma Correlates with Poor Prognosis