Ann Lab Med.
2013 Nov;33(6):431-440. 10.3343/alm.2013.33.6.431.
Comparison of Methylation Profiling in Cancerous and Their Corresponding Normal Tissues from Korean Patients with Breast Cancer
- Affiliations
-
- 1Division of Surgical Oncology, Department of Surgery, Gyeongsang National University Hospital, Jinju, Korea.
- 2Department of Laboratory Medicine, Biomedical Research Institute, Pusan National University Hospital, Busan, Korea. iskim0710@gmail.com
- 3Department of Laboratory Medicine, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 4Department of Pathology, Gyeongsang National University Hospital, Jinju, Korea.
- KMID: 1711317
- DOI: http://doi.org/10.3343/alm.2013.33.6.431
Abstract
- BACKGROUND
Aberrant DNA hypermethylation plays a pivotal role in carcinogenesis and disease progression; therefore, accurate measurement of differential gene methylation patterns among many genes is likely to reveal biomarkers for improved risk assessment. We evaluated the gene hypermethylation profiles of primary breast tumors and their corresponding normal tissues and investigated the association between major clinicopathological features and gene hypermethylation.
METHODS
A single reaction using methylation-specific multiplex ligation-dependent probe amplification was used to analyze the DNA methylation status of 24 tumor suppressor genes in 60 cancerous tissues and their corresponding normal tissues from patients with primary breast cancer.
RESULTS
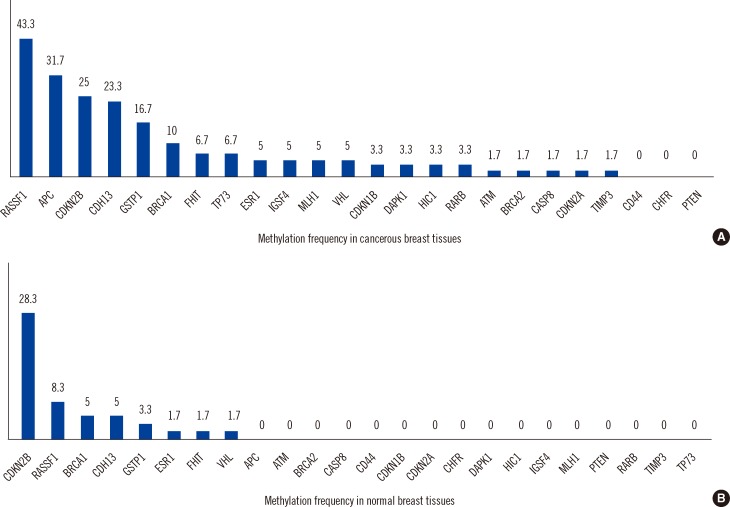
In cancerous breast tissues, 21 of 24 genes displayed promoter methylation in one or more samples. The most frequently methylated genes included RASSF1 (43.3%), APC (31.7%), CDKN2B (25.0%), CDH13 (23.3%), GSTP1 (16.7%), and BRCA1 (10%). APC was associated with lymph node metastasis, and BRCA1 was associated with negative estrogen receptor and negative progesterone receptor expression. In normal breast tissues, 8 of 24 tumor suppressor genes displayed promoter hypermethylation; CDKN2B (28.3%) and RASSF1 (8.3%) hypermethylation were most frequently observed.
CONCLUSIONS
RASSF1 and CDKN2B hypermethylation in Korean breast cancer patients were the most frequent in cancerous tissue and corresponding normal tissue, respectively. Our data indicates that methylation of specific genes is a frequent event in morphologically normal breast tissues adjacent to breast tumors as well as the corresponding breast cancers. This study also suggests that gene methylation is linked to various pathological features of breast cancer; however, this requires confirmation in a larger study.
Keyword
MeSH Terms
-
Adult
Breast/metabolism
Breast Neoplasms/*genetics/metabolism/pathology
Cyclin-Dependent Kinase Inhibitor p15/genetics
*DNA Methylation
Female
Humans
Lymphatic Metastasis
Middle Aged
Promoter Regions, Genetic
Republic of Korea
Tumor Suppressor Proteins/genetics
Cyclin-Dependent Kinase Inhibitor p15
Tumor Suppressor Proteins
Figure
Reference
-
1. The Korea Central Cancer Registry. Annual report of cancer statistics in Korea in 2009. Seoul: The Korean Ministry of Health and Welfare;2011.2. Li S, Rong M, Iacopetta B. DNA hypermethylation in breast cancer and its association with clinicopathological features. Cancer Lett. 2006; 237:272–280. PMID: 16029926.
Article3. Van De Voorde L, Speeckaert R, Van Gestel D, Bracke M, De Neve W, Delanghe J, et al. DNA methylation-based biomarkers in serum of patients with breast cancer. Mutat Res. 2012; 751:304–325. PMID: 22698615.
Article4. Marzese DM, Hoon DS, Chong KK, Gago FE, Orozco JI, Tello OM, et al. DNA methylation index and methylation profile of invasive ductal breast tumors. J Mol Diagn. 2012; 14:613–622. PMID: 22925694.
Article5. Cheol Kim D, Thorat MA, Lee MR, Cho SH, Vasiljevíc N, Scibior-Bentkowska D, et al. Quantitative DNA methylation and recurrence of breast cancer: a study of 30 candidate genes. Cancer Biomark. 2012; 11:75–88. PMID: 23011154.
Article6. Zhu W, Qin W, Hewett JE, Sauter ER. Quantitative evaluation of DNA hypermethylation in malignant and benign breast tissue and fluids. Int J Cancer. 2010; 126:474–482. PMID: 19618401.
Article7. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002; 21:5427–5440. PMID: 12154405.
Article8. Esteller M. Cancer epigenetics: DNA methylation and chromatin alterations in human cancer. Adv Exp Med Biol. 2003; 532:39–49. PMID: 12908548.
Article9. Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002; 21:5462–5482. PMID: 12154408.
Article10. Xu J, Shetty PB, Feng W, Chenault C, Bast RC Jr, Issa JP, et al. Methylation of HIN-1, RASSF1A, RIL and CDH13 in breast cancer is associated with clinical characteristics, but only RASSF1A methylation is associated with outcome. BMC Cancer. 2012; 12:243. PMID: 22695491.
Article11. Wang S, Dorsey TH, Terunuma A, Kittles RA, Ambs S, Kwabi-Addo B. Relationship between tumor DNA methylation status and patient characteristics in African-American and European-American women with breast cancer. PLoS One. 2012; 7:e37928. PMID: 22701537.
Article12. van Hoesel AQ, van de Velde CJ, Kuppen PJ, Putter H, de Kruijf EM, van Nes JG, et al. Primary tumor classification according to methylation pattern is prognostic in patients with early stage ER-negative breast cancer. Breast Cancer Res Treat. 2012; 131:859–869. PMID: 21479925.
Article13. Tserga A, Michalopoulos NV, Levidou G, Korkolopoulou P, Zografos G, Patsouris E, et al. Association of aberrant DNA methylation with clinicopathological features in breast cancer. Oncol Rep. 2012; 27:1630–1638. PMID: 22159596.
Article14. Szyf M. DNA methylation signatures for breast cancer classification and prognosis. Genome Med. 2012; 4:26. PMID: 22494847.
Article15. Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002; 41:154–161. PMID: 12405947.
Article16. Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007; 25:118–145. PMID: 17159189.
Article17. Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005; 33:e128. PMID: 16106041.
Article18. Van der Auwera I, Bovie C, Svensson C, Trinh XB, Limame R, van Dam P, et al. Quantitative methylation profiling in tumor and matched morphologically normal tissues from breast cancer patients. BMC Cancer. 2010; 10:97. PMID: 20226036.
Article19. Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, et al. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011; 458:73–84. PMID: 21120523.
Article20. Kim JH, Shin MH, Kweon SS, Park MH, Yoon JH, Lee JS, et al. Evaluation of promoter hypermethylation detection in serum as a diagnostic tool for breast carcinoma in Korean women. Gynecol Oncol. 2010; 118:176–181. PMID: 20466412.
Article21. Marzese DM, Gago FE, Vargas-Roig LM, Roqué M. Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes. 2010; 24:271–280. PMID: 20561583.22. Buyru N, Altinisik J, Ozdemir F, Demokan S, Dalay N. Methylation profiles in breast cancer. Cancer Invest. 2009; 27:307–312. PMID: 19194828.
Article23. Marzese DM, Gago FE, Vargas-Roig LM, Roque M. Simultaneous analysis of the methylation profile of 26 cancer related regions in invasive breast carcinomas by MS-MLPA and drMS-MLPA. Mol Cell Probes. 2010; 24:271–280. PMID: 20561583.24. Dammann R, Yang G, Pfeifer GP. Hypermethylation of the cpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 2001; 61:3105–3109. PMID: 11306494.25. Honorio S, Agathanggelou A, Schuermann M, Pankow W, Viacava P, Maher ER, et al. Detection of RASSF1A aberrant promoter hypermethylation in sputum from chronic smokers and ductal carcinoma in situ from breast cancer patients. Oncogene. 2003; 22:147–150. PMID: 12527916.
Article26. Yeo W, Wong WL, Wong N, Law BK, Tse GM, Zhong S. High frequency of promoter hypermethylation of RASSF1A in tumorous and non-tumourous tissue of breast cancer. Pathology. 2005; 37:125–130. PMID: 16028839.
Article27. Müller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003; 63:7641–7645. PMID: 14633683.28. Mehrotra J, Vali M, McVeigh M, Kominsky SL, Fackler MJ, Lahti-Domenici J, et al. Very high frequency of hypermethylated genes in breast cancer metastasis to the bone, brain, and lung. Clin Cancer Res. 2004; 10:3104–3109. PMID: 15131050.
Article29. Dodge JE, Munson C, List AF. KG-1 and KG-1a model the p15 CpG island methylation observed in acute myeloid leukemia patients. Leuk Res. 2001; 25:917–925. PMID: 11532526.
Article30. Aggerholm A, Guldberg P, Hokland M, Hokland P. Extensive intra- and interindividual heterogeneity of p15INK4B methylation in acute myeloid leukemia. Cancer Res. 1999; 59:436–441. PMID: 9927059.31. Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996; 56:722–727. PMID: 8631003.32. Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003; 17:1813–1819. PMID: 12970781.
Article33. Qin Y, Liu JY, Li B, Sun ZL, Sun ZF. Association of low p16INK4a and p15INK4b mRNAs expression with their CpG islands methylation with human hepatocellular carcinogenesis. World J Gastroenterol. 2004; 10:1276–1280. PMID: 15112341.
Article34. Ishiguro A, Takahata T, Saito M, Yoshiya G, Tamura Y, Sasaki M, et al. Influence of methylated p15 and p16 genes on clinicopathological features in colorectal cancer. J Gastroenterol Hepatol. 2006; 21:1334–1339. PMID: 16872319.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Quantitative Analysis of Cancer-associated Gene Methylation Connected to Risk Factors in Korean Colorectal Cancer Patients
- Chromosomal Losses are Associated with Hypomethylation of the Gene-Control Regions in the Stomach with a Low Number of Active Genes
- Promoter Methylation Profiles and Its Association with Clinicopathological Features in Breast Cancer
- The Characterization of CpG Methylation of ERalpha and ERbeta Gene in the Breast Cancer
- Methylation Patterns of Tumor Suppressor Genes in Breast DCIS Tumors