Ann Lab Med.
2012 Jul;32(4):307-311. 10.3343/alm.2012.32.4.307.
Ring Chromosome 5 in Acute Myeloid Leukemia Defined by Whole-genome Single Nucleotide Polymorphism Array
- Affiliations
-
- 1Department of Laboratory Medicine, Ewha Womans University School of Medicine, Seoul, Korea. JungWonH@ewha.ac.kr
- 2Department of Internal Medicine, Ewha Womans University School of Medicine, Seoul, Korea. cmseong@ewha.ac.kr
- KMID: 1380093
- DOI: http://doi.org/10.3343/alm.2012.32.4.307
Abstract
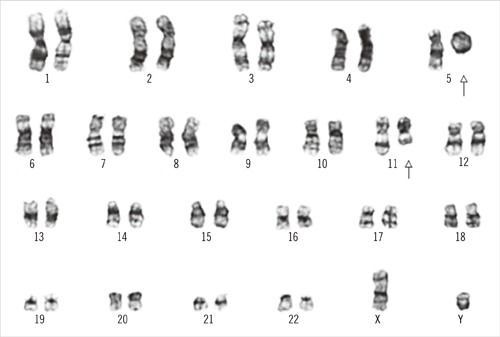
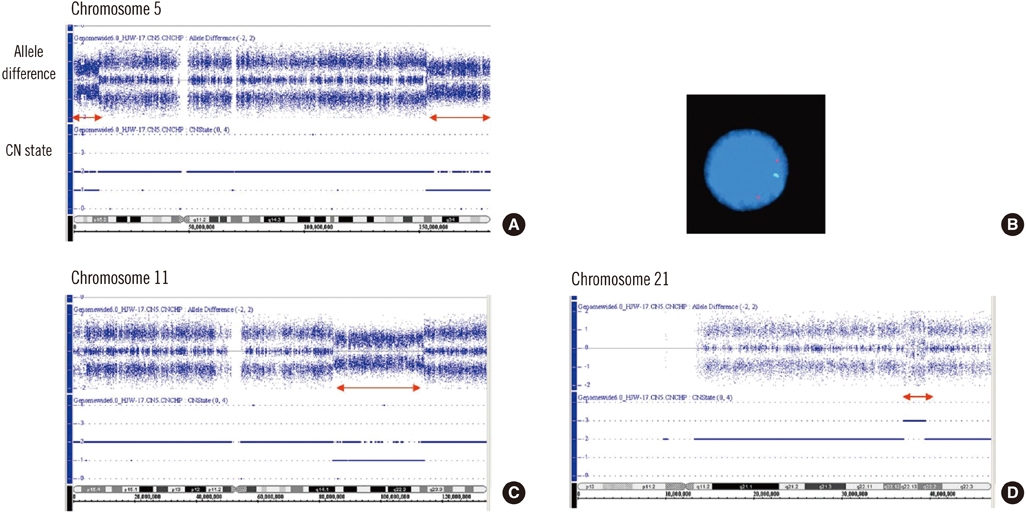
- Chromosomes forming a corresponding ring cannot be clearly defined by conventional cytogenetics or FISH. Karyotypic analyses using whole-genome single nucleotide polymorphism arrays (SNP-A) may result in the identification of previously cryptic lesions and allow for more precise definition of breakpoints. We describe a case of AML with metaphase cells bearing -5, del(11)(q22), and +r. With SNP-A, a 5p-terminal deletion (11 megabases [Mb]), a 5q-terminal deletion (27 Mb), an 11q-interstitial deletion (29 Mb), and a 21q gain (3 Mb) were identified. Therefore, the G-banded karyotype was revised as 46, XY, r(5)(p15. 2q33.2), del(11)(q14.1q23.2), dup(21)(q22.13q22.2)[18]/46,XY[2]. SNP-A could be a powerful tool for characterizing ring chromosomes in which the involved chromosomes or bands cannot be precisely identified by conventional cytogenetics or FISH.
Keyword
MeSH Terms
Figure
Cited by 1 articles
-
Abnormalities in Chromosomes 5 and 7 in Myelodysplastic Syndrome and Acute Myeloid Leukemia
Tulene S. Kendrick, Daria Buic, Kathy A. Fuller, Wendy N. Erber
Ann Lab Med. 2025;45(2):133-145. doi: 10.3343/alm.2024.0477.
Reference
-
1. Dessen P, Huret JL, editors. Chromosome. Atlas genet cytogenet oncol haematol. Updated on Dec 2011. http://AtlasGeneticsOncology.org/Indexbychrom/idxa_5.html.2. Mitelman F, Johansson B, Mertens F, editors. Mitelman database of chromosome aberrations and gene fusions in cancer. Updated on Nov 2011. http://cgap.nci.nih.gov/Chromosomes/Mitelman.3. Herry A, Douet-Guilbert N, Morel F, Le Bris MJ, De Braekeleer M. Redefining monosomy 5 by molecular cytogenetics in 23 patients with MDS/AML. Eur J Haematol. 2007. 78:457–467.
Article4. Gebhart E. Ring chromosomes in human neoplasias. Cytogenet Genome Res. 2008. 121:149–173.
Article5. Gisselsson D, Höglund M, Mertens F, Johnsson B, Dal Cin P, Van den Berghe H, et al. The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells. Hum Genet. 1999. 104:315–325.
Article6. Maciejewski JP, Tiu RV, O'Keefe C. Application of array-based whole genome scanning technologies as a cytogenetic tool in haematological malignancies. Br J Haematol. 2009. 146:479–488.
Article7. Dutt A, Beroukhim R. Single nucleotide polymorphism array analysis of cancer. Curr Opin Oncol. 2007. 19:43–49.
Article8. Shaffer LG, Slovak ML, editors. ISCN 2009: an international system for human cytogenetic nomenclature. 2009. Basel: Karger.9. Makishima H, Maciejewski JP. Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res. 2011. 17:3913–3923.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Additional Genomic Aberrations Identified by Single Nucleotide Polymorphism Array-Based Karyotyping in an Acute Myeloid Leukemia Case with Isolated del(20q) Abnormality
- Clinical Significance of Previously Cryptic Copy Number Alterations and Loss of Heterozygosity in Pediatric Acute Myeloid Leukemia and Myelodysplastic Syndrome Determined Using Combined Array Comparative Genomic Hybridization plus Single-Nucleotide Polymorphism Microarray Analyses
- A Case of Acute Myeloid Leukemia with Bone Marrow Basophilia and Dysmegakaryocytic Hyperplasia with Isochromosome 17q as a Sole Cytogenetic Abnormality: A Clinical Study with Literature Review
- Comparison of the Affymetrix SNP Array 5.0 and Oligoarray Platforms for Defining CNV
- Effects of Somatic Mutations Are Associated with SNP in the Progression of Individual Acute Myeloid Leukemia Patient: The Two-Hit Theory Explains Inherited Predisposition to Pathogenesis