Korean Circ J.
2023 Jan;53(1):1-16. 10.4070/kcj.2022.0295.
Strategy of Patient-Specific Therapeutics in Cardiovascular Disease Through Single-Cell RNA Sequencing
- Affiliations
-
- 1Strategic Center of Cell and Bio Therapy for Heart, Diabetes & Cancer, Biomedical Research Institute, Seoul National University Hospital, Seoul, Korea
- 2Department of Medicine, Seoul National University College of Medicine, Seoul National University, Seoul, Korea
- KMID: 2538101
- DOI: http://doi.org/10.4070/kcj.2022.0295
Abstract
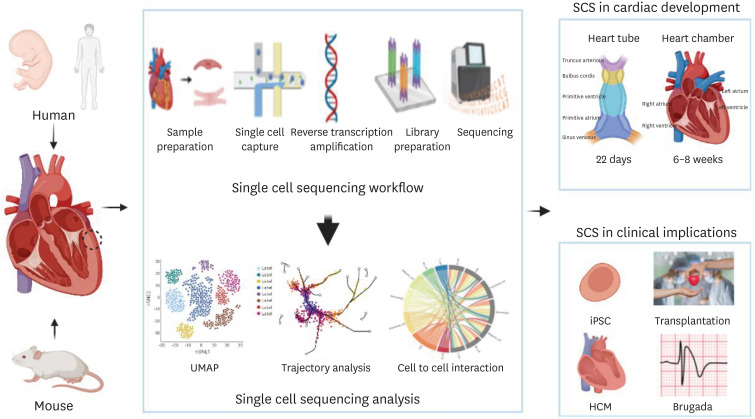
- Recently, single cell RNA sequencing (scRNA-seq) technology has enabled the discovery of novel or rare subtypes of cells and their characteristics. This technique has advanced unprecedented biomedical research by enabling the profiling and analysis of the transcriptomes of single cells at high resolution and throughput. Thus, scRNA-seq has contributed to recent advances in cardiovascular research by the generation of cell atlases of heart and blood vessels and the elucidation of mechanisms involved in cardiovascular development and diseases. This review summarizes the overall workflow of the scRNA-seq technique itself and key findings in the cardiovascular development and diseases based on the previous studies. In particular, we focused on how the single-cell sequencing technology can be utilized in clinical field and precision medicine to treat specific diseases.
Figure
Reference
-
1. Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018; 14:479–492. PMID: 29789704.
Article2. Paik DT, Cho S, Tian L, Chang HY, Wu JC. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020; 17:457–473. PMID: 32231331.
Article3. Pimpalwar N, Czuba T, Smith ML, Nilsson J, Gidlöf O, Smith JG. Methods for isolation and transcriptional profiling of individual cells from the human heart. Heliyon (Lond). 2020; 6:e05810.
Article4. Yamada S, Nomura S. Review of single-cell RNA sequencing in the heart. Int J Mol Sci. 2020; 21:8345. PMID: 33172208.
Article5. Choi YH, Kim JK. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol Cells. 2019; 42:189–199. PMID: 30764602.6. Hulin A, Hortells L, Gomez-Stallons MV, et al. Maturation of heart valve cell populations during postnatal remodeling. Development. 2019; 146:dev173047. PMID: 30796046.
Article7. Gladka MM, Molenaar B, de Ruiter H, et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018; 138:166–180. PMID: 29386203.
Article8. Panther F, Williams T, Ritter O. Inhibition of the calcineurin-NFAT signalling cascade in the treatment of heart failure. Recent Patents Cardiovasc Drug Discov. 2009; 4:180–186.
Article9. Vidal R, Wagner JUG, Braeuning C, et al. Transcriptional heterogeneity of fibroblasts is a hallmark of the aging heart. JCI Insight. 2019; 4:e131092. PMID: 31723062.10. Gladka MM. Single-cell RNA sequencing of the adult mammalian heart-state-of-the-art and future perspectives. Curr Heart Fail Rep. 2021; 18:64–70. PMID: 33629280.
Article11. Ramilowski JA, Goldberg T, Harshbarger J, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015; 6:7866. PMID: 26198319.12. Skelly DA, Squiers GT, McLellan MA, et al. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Reports. 2018; 22:600–610. PMID: 29346760.13. Farbehi N, Patrick R, Dorison A, et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife. 2019; 8:e43882. PMID: 30912746.14. Ren Z, Yu P, Li D, et al. Single-cell reconstruction of progression trajectory reveals intervention principles in pathological cardiac hypertrophy. Circulation. 2020; 141:1704–1719. PMID: 32098504.15. Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods. 2014; 11:163–166. PMID: 24363023.
Article16. Deng Q, Ramsköld D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014; 343:193–196. PMID: 24408435.
Article17. Wang M, Gu M, Liu L, Liu Y, Tian L. Single-cell RNA sequencing (scRNA-seq) in cardiac tissue: applications and limitations. Vasc Health Risk Manag. 2021; 17:641–657. PMID: 34629873.18. Betts JG, Young KA, Wise JA, et al. 2013.19. Samad T, Wu SM. Single cell RNA sequencing approaches to cardiac development and congenital heart disease. Semin Cell Dev Biol. 2021; 118:129–135. PMID: 34006454.
Article20. Chen Z, Wei L, Duru F, Chen L. Single-cell RNA sequencing: in-depth decoding of heart biology and cardiovascular diseases. Curr Genomics. 2020; 21:585–601. PMID: 33414680.
Article21. Litviňuková M, Talavera-López C, Maatz H, et al. Cells of the adult human heart. Nature. 2020; 588:466–472. PMID: 32971526.
Article22. Cui Y, Zheng Y, Liu X, et al. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep. 2019; 26:1934–1950.e5. PMID: 30759401.23. Liu X, Chen W, Li W, et al. Single-cell RNA-seq of the developing cardiac outflow tract reveals convergent development of the vascular smooth muscle cells. Cell Rep. 2019; 28:1346–1361.e4. PMID: 31365875.
Article24. Xiong H, Luo Y, Yue Y, et al. Single-cell transcriptomics reveals chemotaxis-mediated intraorgan crosstalk during cardiogenesis. Circ Res. 2019; 125:398–410. PMID: 31221018.
Article25. DeLaughter DM, Bick AG, Wakimoto H, et al. Single-cell resolution of temporal gene expression during heart development. Dev Cell. 2016; 39:480–490. PMID: 27840107.
Article26. Li G, Xu A, Sim S, et al. Transcriptomic profiling maps anatomically patterned subpopulations among single embryonic cardiac cells. Dev Cell. 2016; 39:491–507. PMID: 27840109.
Article27. Lescroart F, Wang X, Lin X, et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science. 2018; 359:1177–1181. PMID: 29371425.28. Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014; 9:171–181. PMID: 24385147.29. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006; 126:663–676. PMID: 16904174.
Article30. Friedman CE, Nguyen Q, Lukowski SW, et al. Single-cell transcriptomic analysis of cardiac differentiation from human PSCs reveals HOPX-dependent cardiomyocyte maturation. Cell Stem Cell. 2018; 23:586–598.e8. PMID: 30290179.
Article31. Churko JM, Garg P, Treutlein B, et al. Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis. Nat Commun. 2018; 9:4906. PMID: 30464173.32. Yap L, Wang JW, Moreno-Moral A, et al. In vivo generation of post-infarct human cardiac muscle by laminin-promoted cardiovascular progenitors. Cell Rep. 2019; 26:3231–3245.e9. PMID: 30893597.33. Wehrens M, de Leeuw AE, Wright-Clark M, et al. Single-cell transcriptomics provides insights into hypertrophic cardiomyopathy. Cell Rep. 2022; 39:110809. PMID: 35545053.
Article34. Kopecky BJ, Dun H, Amrute JM, et al. Donor macrophages modulate rejection after heart transplantation. Circulation. 2022; 146:623–638. PMID: 35880523.
Article35. Chang Y, Li X, Cheng Q, et al. Single-cell transcriptomic identified HIF1A as a target for attenuating acute rejection after heart transplantation. Basic Res Cardiol. 2021; 116:64. PMID: 34870762.36. Tambi R, Abdel Hameid R, Bankapur A, et al. Single-cell transcriptomics trajectory and molecular convergence of clinically relevant mutations in Brugada syndrome. Am J Physiol Heart Circ Physiol. 2021; 320:H1935–H1948. PMID: 33797273.37. Bui TV, Hwang JW, Lee JH, Park HJ, Ban K. Challenges and limitations of strategies to promote therapeutic potential of human mesenchymal stem cells for cell-based cardiac repair. Korean Circ J. 2021; 51:97–113. PMID: 33525065.38. Almeida SO, Skelton RJ, Adigopula S, Ardehali R. Arrhythmia in stem cell transplantation. Card Electrophysiol Clin. 2015; 7:357–370. PMID: 26002399.39. Kim KH, Pereira NL. Genetics of cardiomyopathy: clinical and mechanistic implications for heart failure. Korean Circ J. 2021; 51:797–836. PMID: 34327881.40. Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002; 287:1308–1320. PMID: 11886323.41. Bajpai G, Schneider C, Wong N, et al. The human heart contains distinct macrophage subsets with divergent origins and functions. Nat Med. 2018; 24:1234–1245. PMID: 29892064.42. Ortega A, Roselló-Lletí E, Tarazón E, et al. Endoplasmic reticulum stress induces different molecular structural alterations in human dilated and ischemic cardiomyopathy. PLoS One. 2014; 9:e107635. PMID: 25226522.43. Fan P, Zhang L, Cheng T, et al. MiR-590-5p inhibits pathological hypertrophy mediated heart failure by targeting RTN4. J Mol Histol. 2021; 52:955–964. PMID: 34406553.44. Wu H, Malone AF, Donnelly EL, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. 2018; 29:2069–2080. PMID: 29980650.45. Ferguson A, Iasella C, Chen W, et al. Gene expression profiling of lung transplant patients using next-generation sequencing to identify biomarkers for chronic lung allograft dysfunction. Am J Respir Crit Care Med. 2018; 197:A4732.46. Lavine KJ, Epelman S, Uchida K, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014; 111:16029–16034. PMID: 25349429.
Article47. Talib AK, Nogami A. Catheter ablation for Brugada syndrome. Korean Circ J. 2020; 50:289–301. PMID: 31960637.48. Cao J, O’Day DR, Pliner HA, et al. A human cell atlas of fetal gene expression. Science. 2020; 370:eaba7721. PMID: 33184181.49. Tucker NR, Chaffin M, Fleming SJ, et al. Transcriptional and cellular diversity of the human heart. Circulation. 2020; 142:466–482. PMID: 32403949.
Article51. Aspinall MG, Hamermesh RG. Realizing the promise of personalized medicine. Harv Bus Rev. 2007; 85:108–117. 165PMID: 17972499.52. Vicente AM, Ballensiefen W, Jönsson JI. How personalised medicine will transform healthcare by 2030: the ICPerMed vision. J Transl Med. 2020; 18:180. PMID: 32345312.
Article53. Atkinson M. Mapping the 100 trillion cells that make up your body. News-University of Florida [Internet]. Gainesville (FL): University of Florida;2018. cited 2022 October 24. Available from: https://news.ufl.edu/articles/2018/10/mapping-the-100-trillion-cells-that-make-up-your-body.html.54. Marusyk A, Janiszewska M, Polyak K. Intratumor heterogeneity: the Rosetta stone of therapy resistance. Cancer Cell. 2020; 37:471–484. PMID: 32289271.
Article55. Lee MC, Lopez-Diaz FJ, Khan SY, et al. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc Natl Acad Sci U S A. 2014; 111:E4726–E4735. PMID: 25339441.56. Spiller W, Jung KJ, Lee JY, Jee SH. Precision medicine and cardiovascular health: insights from mendelian randomization analyses. Korean Circ J. 2020; 50:91–111. PMID: 31845553.
Article57. Leopold JA, Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res. 2018; 122:1302–1315. PMID: 29700074.
Article58. HuBMAP Consortium. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature. 2019; 574:187–192. PMID: 31597973.59. Brandenburg RO. Report of the WHO/ISFC task force on the definition and classification of cardiomyopathies. Br Heart J. 1980; 44:672–673. PMID: 7459150.60. Elliott PM. Personalized medicine for dilated cardiomyopathy. Eur Heart J. 2021; 42:175–177. PMID: 33175136.
Article61. Reichart D, Lindberg EL, Maatz H, et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies. Science. 2022; 377:eabo1984. PMID: 35926050.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Perspectives on single-nucleus RNA sequencing in different cell types and tissues
- Single cell RNA sequencing used in asthma research
- A semi-automatic cell type annotation method for single-cell RNA sequencing dataset
- Single Cell RNA-Sequencing for the Study of Atherosclerosis
- Genetics of Alzheimer's Disease