Cancer Res Treat.
2023 Jan;55(1):28-40. 10.4143/crt.2021.1572.
Interlaboratory Comparison Study (Ring Test) of Next-Generation Sequencing–Based NTRK Fusion Detection in South Korea
- Affiliations
-
- 1Department of Pathology, Konkuk University Medical Center, Konkuk University School of Medicine, Seoul, Korea
- 2Department of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea
- 3Department of Pathology, Seoul National University Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea
- 4Department of Pathology, Yonsei University College of Medicine, Seoul, Korea
- 5Department of Hospital Pathology, Seoul St. Mary's Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 6Department of Pathology, Asan Medical Center, University of Ulsan College of Medicine, Asan Medical Center, Seoul, Korea
- 7Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- KMID: 2537989
- DOI: http://doi.org/10.4143/crt.2021.1572
Abstract
- Purpose
Tropomyosin receptor kinase (TRK) inhibitors are approved for the treatment of neurotrophic receptor tyrosine kinase (NTRK) fusion-positive tumors. The detection of NTRK fusion using a validated method is required before therapeutic application. An interlaboratory comparison study of next-generation sequencing (NGS)–based NTRK gene fusion detection with validated clinical samples was conducted at six major hospitals in South Korea.
Materials and Methods
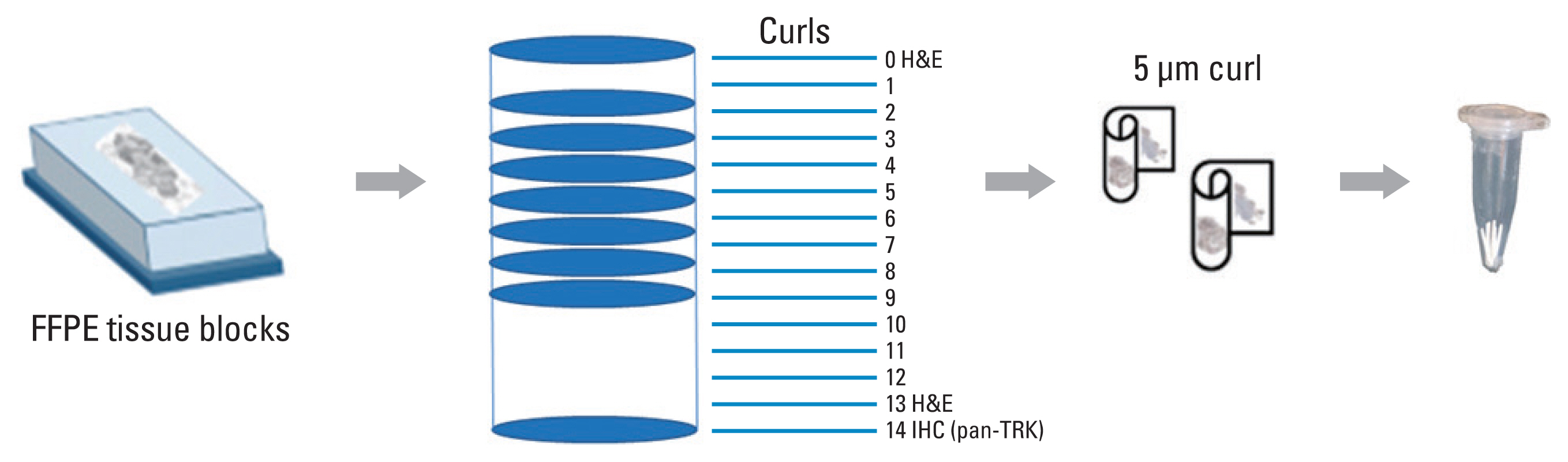
A total of 18 samples, including a positive standard reference and eight positive and nine negative clinical samples, were validated using the VENTANA pan-TRK (EPR17341) and TruSight Oncology 500 assays. These samples were then tested using four different NGS panels currently being used at the six participating institutions.
Results
NTRK fusions were not detected in any of the nine negative clinical samples, demonstrating 100% specificity in all six participating institutions. All assays showed 100% analytical sensitivity to identify the NTRK fusion status in formalin-fixed paraffin-embedded (FFPE) samples, although with variable clinical sensitivity. False-negative results were due to low tumor purity, poor RNA quality, and DNA-based sequencing panel. The RNA-based targeted NGS assay showed an overall high success rate of identifying NTRK fusion status in FFPE samples.
Conclusion
This study is the first to test the proficiency of NGS-based NTRK detection in South Korea with the largest participating institutions. RNA-based NGS assays to detect NTRK fusions can accurately characterize fusion transcripts if sufficient RNA of adequate quality is available. The comparative performance data will support the implementation of targeted NGS-based sequencing assays for NTRK fusion detection in routine diagnostics.
Keyword
Figure
Cited by 1 articles
-
Validation and Clinical Application of ONCOaccuPanel for Targeted Next-Generation Sequencing of Solid Tumors
Moonsik Kim, Changseon Lee, Juyeon Hong, Juhee Kim, Ji Yun Jeong, Nora Jee-Young Park, Ji-Eun Kim, Ji Young Park
Cancer Res Treat. 2022;55(2):429-441. doi: 10.4143/crt.2022.891.
Reference
-
References
1. Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015; 5:25–34.2. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018; 15:731–47.
Article3. Okamura R, Boichard A, Kato S, Sicklick JK, Bazhenova L, Kurzrock R. Analysis of NTRK alterations in pan-cancer adult and pediatric malignancies: implications for NTRK-targeted therapeutics. JCO Precis Oncol. 2018; 2018:PO.1800183.4. Federman N, McDermott R. Larotrectinib, a highly selective tropomyosin receptor kinase (TRK) inhibitor for the treatment of TRK fusion cancer. Expert Rev Clin Pharmacol. 2019; 12:931–9.
Article5. Scott LJ. Larotrectinib: first global approval. Drugs. 2019; 79:201–6.
Article6. Chen Y, Chi P. Basket trial of TRK inhibitors demonstrates efficacy in TRK fusion-positive cancers. J Hematol Oncol. 2018; 11:78.
Article7. Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, et al. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1–2 trials. Lancet Oncol. 2020; 21:271–82.
Article8. Gatalica Z, Xiu J, Swensen J, Vranic S. Molecular characterization of cancers with NTRK gene fusions. Mod Pathol. 2019; 32:147–53.
Article9. Hsiao SJ, Zehir A, Sireci AN, Aisner DL. Detection of tumor NTRK gene fusions to identify patients who may benefit from tyrosine kinase (TRK) inhibitor therapy. J Mol Diagn. 2019; 21:553–71.
Article10. FDA approves companion diagnostic to identify NTRK fusions in solid tumors for Vitrakvi [Internet]. Silver Spring MD: U.S. Food and Drug Administration;2020. [cited 2021 Dec 10]. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-companion-diagnostic-identify-ntrk-fusions-solid-tumors-vitrakvi .11. Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk immunohistochemistry is an efficient and reliable screen for the detection of NTRK fusions. Am J Surg Pathol. 2017; 41:1547–51.
Article12. Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK fusion detection across multiple assays and 33,997 cases: diagnostic implications and pitfalls. Mod Pathol. 2020; 33:38–46.
Article13. Penault-Llorca F, Rudzinski ER, Sepulveda AR. Testing algorithm for identification of patients with TRK fusion cancer. J Clin Pathol. 2019; 72:460–7.
Article14. Xu B, Haroon Al Rasheed MR, Antonescu CR, Alex D, Frosina D, Ghossein R, et al. Pan-Trk immunohistochemistry is a sensitive and specific ancillary tool for diagnosing secretory carcinoma of the salivary gland and detecting ETV6-NTRK3 fusion. Histopathology. 2020; 76:375–82.
Article15. Bourgeois JM, Knezevich SR, Mathers JA, Sorensen PH. Molecular detection of the ETV6-NTRK3 gene fusion differentiates congenital fibrosarcoma from other childhood spindle cell tumors. Am J Surg Pathol. 2000; 24:937–46.
Article16. Davis JL, Lockwood CM, Albert CM, Tsuchiya K, Hawkins DS, Rudzinski ER. Infantile NTRK-associated mesenchymal tumors. Pediatr Dev Pathol. 2018; 21:68–78.17. Rubin BP, Chen CJ, Morgan TW, Xiao S, Grier HE, Kozakewich HP, et al. Congenital mesoblastic nephroma t(12;15) is associated with ETV6-NTRK3 gene fusion: cytogenetic and molecular relationship to congenital (infantile) fibrosarcoma. Am J Pathol. 1998; 153:1451–8.18. Skalova A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010; 34:599–608.
Article19. Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002; 2:367–76.
Article20. Benayed R, Offin M, Mullaney K, Sukhadia P, Rios K, Desmeules P, et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019; 25:4712–22.
Article21. Solomon JP, Hechtman JF. Detection of NTRK fusions: merits and limitations of current diagnostic platforms. Cancer Res. 2019; 79:3163–8.22. Haynes BC, Blidner RA, Cardwell RD, Zeigler R, Gokul S, Thibert JR, et al. An integrated next-generation sequencing system for analyzing DNA mutations, gene fusions, and RNA expression in lung cancer. Transl Oncol. 2019; 12:836–45.23. Pfarr N, Kirchner M, Lehmann U, Leichsenring J, Merkelbach-Bruse S, Glade J, et al. Testing NTRK testing: Wet-lab and in silico comparison of RNA-based targeted sequencing assays. Genes Chromosomes Cancer. 2020; 59:178–88.24. von Ahlfen S, Missel A, Bendrat K, Schlumpberger M. Determinants of RNA quality from FFPE samples. PLoS One. 2007; 2:e1261.25. Kirchner M, Neumann O, Volckmar AL, Stogbauer F, Allgauer M, Kazdal D, et al. RNA-based detection of gene fusions in formalin-fixed and paraffin-embedded solid cancer samples. Cancers (Basel). 2019; 11:1309.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Development of an RNA sequencing panel to detect gene fusions in thyroid cancer
- Performance Evaluation of BRCA1/2 Genetic Test Using Next-Generation Sequencing Based on Target Capture Method
- Korean Society for Genetic Diagnostics Guidelines for Validation of Next-Generation Sequencing-Based Somatic Variant Detection in Hematologic Malignancies
- Detection of Mosaic Sequence Variants Associated with Human Genetic Diseases
- Recent Advances in the Clinical Application of Next-Generation Sequencing