Korean J Pain.
2023 Jan;36(1):60-71. 10.3344/kjp.22277.
A positive feedback loop of heparanase/ syndecan1/nerve growth factor regulates cancer pain progression
- Affiliations
-
- 1Department of Anesthesiology, Suqian First People’s Hospital, Suqian City, Jiangsu Province, China
- 2Cancer Institute, The Second Affiliated Hospital of Xuzhou Medical University, Xuzhou City, Jiangsu Province, China
- 3Department of Anesthesiology, Tai’an Central Hospital, Tai’an City, Shandong Province, China
- 4Department of Anesthesiology, Graduate School of Xuzhou Medical University, Xuzhou City, Jiangsu Province, China
- 5Department of Anesthesiology, The Affiliated Hospital of Xuzhou Medical University, Xuzhou City, Jiangsu Province, China
- KMID: 2537613
- DOI: http://doi.org/10.3344/kjp.22277
Abstract
- Background
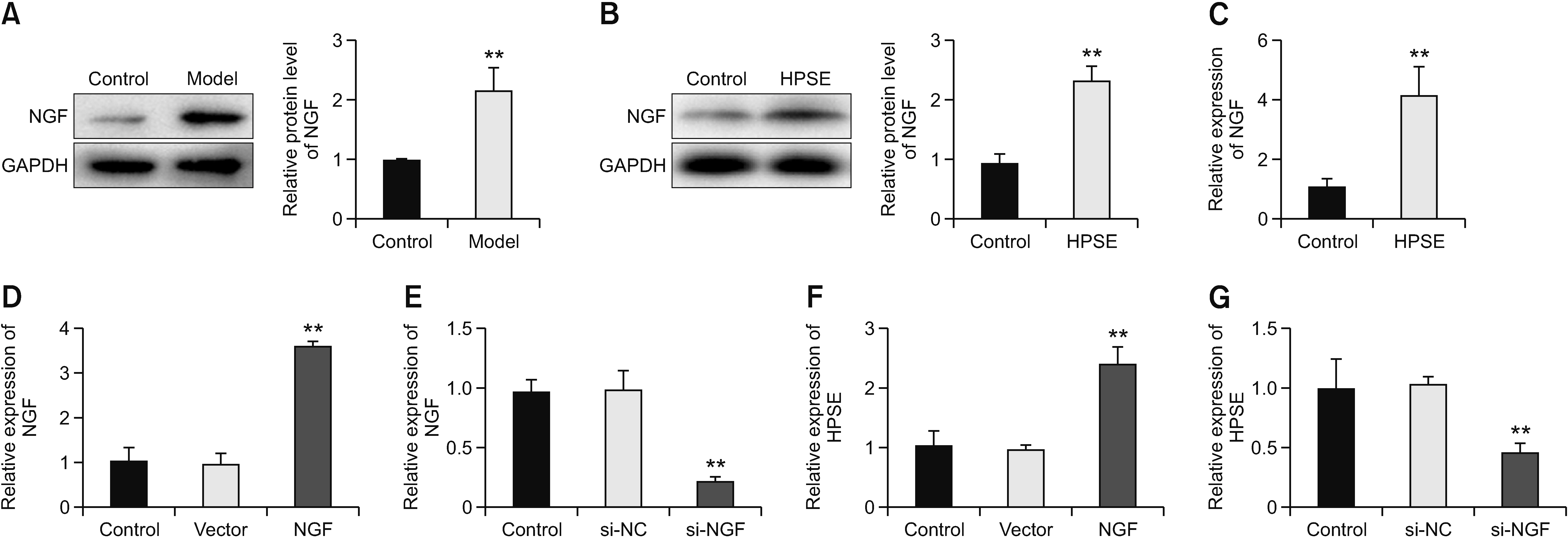
The purpose of this research was to assess the role of heparanase (HPSE)/syndecan1 (SDC1)/nerve growth factor (NGF) on cancer pain from melanoma.
Methods
The influence of HPSE on the biological function of melanoma cells and cancer pain in a mouse model was evaluated. Immunohistochemical staining was used to analyze HPSE and SDC1. HPSE, NGF, and SDC1 were detected using western blot. Inflammatory factors were detected using ELISA assay.
Results
HPSE promoted melanoma cell viability, proliferation, migration, invasion, and tumor growth, as well as cancer pain, while SST0001 treatment reversed the promoting effect of HPSE. HPSE up-regulated NGF, and NGF feedback promoted HPSE. High expression of NGF reversed the inhibitory effect of HPSE down-regulation on melanoma cell phenotype deterioration, including cell viability, proliferation, migration, and invasion. SST0001 down-regulated SDC1 expression. SDC1 reversed the inhibitory effect of SST0001 on cancer pain.
Conclusions
The results showed that HPSE promoted melanoma development and cancer pain by interacting with NGF/SDC1. It provides new insights to better understand the role of HPSE in melanoma and also provides a new direction for cancer pain treatment.
Keyword
Figure
Reference
-
1. Neufeld NJ, Elnahal SM, Alvarez RH. 2017; Cancer pain: a review of epidemiology, clinical quality and value impact. Future Oncol. 13:833–41. DOI: 10.2217/fon-2016-0423. PMID: 27875910.2. Lovell M, Agar M, Luckett T, Davidson PM, Green A, Clayton J. 2013; Australian survey of current practice and guideline use in adult cancer pain assessment and management: perspectives of palliative care physicians. J Palliat Med. 16:1403–9. DOI: 10.1089/jpm.2013.0245. PMID: 24168350. PMCID: PMC3822364.3. Magee D, Bachtold S, Brown M, Farquhar-Smith P. 2019; Cancer pain: where are we now? Pain Manag. 9:63–79. DOI: 10.2217/pmt-2018-0031. PMID: 30516438.4. Sanderson RD, Elkin M, Rapraeger AC, Ilan N, Vlodavsky I. 2017; Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J. 284:42–55. DOI: 10.1111/febs.13932. PMID: 27758044. PMCID: PMC5226874.5. Rivara S, Milazzo FM, Giannini G. 2016; Heparanase: a rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med Chem. 8:647–80. DOI: 10.4155/fmc-2016-0012. PMID: 27057774.6. Doweck I, Feibish N. 2020; Opposing effects of heparanase and heparanase-2 in head & neck cancer. Adv Exp Med Biol. 1221:847–56. DOI: 10.1007/978-3-030-34521-1_37. PMID: 32274741.7. Hermano E, Goldberg R, Rubinstein AM, Sonnenblick A, Maly B, Nahmias D, et al. 2019; Heparanase accelerates obesity-associated breast cancer progression. Cancer Res. 79:5342–54. DOI: 10.1158/0008-5472.CAN-18-4058. PMID: 31481501.8. Heyman B, Yang Y. 2016; Mechanisms of heparanase inhibitors in cancer therapy. Exp Hematol. 44:1002–12. DOI: 10.1016/j.exphem.2016.08.006. PMID: 27576132. PMCID: PMC5083136.9. Jiang X, Tian Y, Xu L, Zhang Q, Wan Y, Qi X, et al. 2019; Inhibition of triple-negative breast cancer tumor growth by electroacupuncture with encircled needling and its mechanisms in a mice xenograft model. Int J Med Sci. 16:1642–51. DOI: 10.7150/ijms.38521. PMID: 31839752. PMCID: PMC6909807.10. Demir IE, Tieftrunk E, Schorn S, Friess H, Ceyhan GO. 2016; Nerve growth factor & TrkA as novel therapeutic targets in cancer. Biochim Biophys Acta. 1866:37–50. DOI: 10.1016/j.bbcan.2016.05.003. PMID: 27264679.11. Wang W, Chen J, Guo X. 2014; The role of nerve growth factor and its receptors in tumorigenesis and cancer pain. Biosci Trends. 8:68–74. DOI: 10.5582/bst.8.68. PMID: 24815383.12. Di Donato M, Cernera G, Migliaccio A, Castoria G. 2019; Nerve growth factor induces proliferation and aggressiveness in prostate cancer cells. Cancers (Basel). 11:784. DOI: 10.3390/cancers11060784. PMID: 31174415. PMCID: PMC6627659.13. Hayakawa Y, Sakitani K, Konishi M, Asfaha S, Niikura R, Tomita H, et al. 2017; Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell. 31:21–34. DOI: 10.1016/j.ccell.2016.11.005. PMID: 27989802. PMCID: PMC5225031.14. Han L, Jiang J, Xue M, Qin T, Xiao Y, Wu E, et al. 2020; Sonic hedgehog signaling pathway promotes pancreatic cancer pain via nerve growth factor. Reg Anesth Pain Med. 45:137–44. DOI: 10.1136/rapm-2019-100991. PMID: 31792027.15. Teixeira FCOB, Götte M. 2020; Involvement of Syndecan-1 and heparanase in cancer and inflammation. Adv Exp Med Biol. 1221:97–135. DOI: 10.1007/978-3-030-34521-1_4. PMID: 32274708.16. Jayatilleke KM, Hulett MD. 2020; Heparanase and the hallmarks of cancer. J Transl Med. 18:453. DOI: 10.1186/s12967-020-02624-1. PMID: 33256730. PMCID: PMC7706218. PMID: 40c9c074652d4073a0d58c5948501b73.17. Sayyad MR, Puchalapalli M, Vergara NG, Wangensteen SM, Moore M, Mu L, et al. 2019; Syndecan-1 facilitates breast cancer metastasis to the brain. Breast Cancer Res Treat. 178:35–49. DOI: 10.1007/s10549-019-05347-0. PMID: 31327090.18. Chute C, Yang X, Meyer K, Yang N, O'Neil K, Kasza I, et al. 2018; Syndecan-1 induction in lung microenvironment supports the establishment of breast tumor metastases. Breast Cancer Res. 20:66. DOI: 10.1186/s13058-018-0995-x. PMID: 29976229. PMCID: PMC6034333.19. Calixto-Campos C, Corrêa MP, Carvalho TT, Zarpelon AC, Hohmann MS, Rossaneis AC, et al. 2015; Quercetin reduces Ehrlich tumor-induced cancer pain in mice. Anal Cell Pathol (Amst). 2015:285708. DOI: 10.1155/2015/285708. PMID: 26351625. PMCID: PMC4550761.20. Pellati F, Borgonetti V, Brighenti V, Biagi M, Benvenuti S, Corsi L. 2018; Cannabis sativa L. and nonpsychoactive cannabinoids: their chemistry and role against oxidative stress, inflammation, and cancer. Biomed Res Int. 2018:1691428. DOI: 10.1155/2018/1691428. PMID: 30627539. PMCID: PMC6304621.21. Masola V, Zaza G, Gambaro G, Franchi M, Onisto M. 2020; Role of heparanase in tumor progression: molecular aspects and therapeutic options. Semin Cancer Biol. 62:86–98. DOI: 10.1016/j.semcancer.2019.07.014. PMID: 31348993.22. Hermano E, Meirovitz A, Meir K, Nussbaum G, Appelbaum L, Peretz T, et al. 2014; Macrophage polarization in pancreatic carcinoma: role of heparanase enzyme. J Natl Cancer Inst. 106:dju332. DOI: 10.1093/jnci/dju332. PMID: 25326645. PMCID: PMC4334800.23. Vlodavsky I, Singh P, Boyango I, Gutter-Kapon L, Elkin M, Sanderson RD, et al. 2016; Heparanase: from basic research to therapeutic applications in cancer and inflammation. Drug Resist Updat. 29:54–75. DOI: 10.1016/j.drup.2016.10.001. PMID: 27912844. PMCID: PMC5447241.24. Ramani VC, Zhan F, He J, Barbieri P, Noseda A, Tricot G, et al. 2016; Targeting heparanase overcomes chemoresistance and diminishes relapse in myeloma. Oncotarget. 7:1598–607. DOI: 10.18632/oncotarget.6408. PMID: 26624982. PMCID: PMC4811483.25. Hao NB, Tang B, Wang GZ, Xie R, Hu CJ, Wang SM, et al. 2015; Hepatocyte growth factor (HGF) upregulates heparanase expression via the PI3K/Akt/NF-κB signaling pathway for gastric cancer metastasis. Cancer Lett. 361:57–66. DOI: 10.1016/j.canlet.2015.02.043. PMID: 25727320.26. Luan Q, Sun J, Li C, Zhang G, Lv Y, Wang G, et al. 2011; Mutual enhancement between heparanase and vascular endothelial growth factor: a novel mechanism for melanoma progression. Cancer Lett. 308:100–11. DOI: 10.1016/j.canlet.2011.04.019. PMID: 21624769.27. Wei RR, Sun DN, Yang H, Yan J, Zhang X, Zheng XL, et al. 2018; CTC clusters induced by heparanase enhance breast cancer metastasis. Acta Pharmacol Sin. 39:1326–37. DOI: 10.1038/aps.2017.189. PMID: 29417941. PMCID: PMC6289387.28. Barash U, Lapidot M, Zohar Y, Loomis C, Moreira A, Feld S, et al. 2018; Involvement of heparanase in the pathogenesis of mesothelioma: basic aspects and clinical applications. J Natl Cancer Inst. 110:1102–14. DOI: 10.1093/jnci/djy032. PMID: 29579286. PMCID: PMC6186523.29. Ramani VC, Vlodavsky I, Ng M, Zhang Y, Barbieri P, Noseda A, et al. 2016; Chemotherapy induces expression and release of heparanase leading to changes associated with an aggressive tumor phenotype. Matrix Biol. 55:22–34. DOI: 10.1016/j.matbio.2016.03.006. PMID: 27016342. PMCID: PMC5033659.30. Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, et al. 2018; β2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. 33:75–90.e7. Erratum in: Cancer Cell 2018; 34: 863-7. DOI: 10.1016/j.ccell.2017.11.007. PMID: 29249692. PMCID: PMC5760435.31. Salvo E, Tu NH, Scheff NN, Dubeykovskaya ZA, Chavan SA, Aouizerat BE, et al. 2021; TNFα promotes oral cancer growth, pain, and Schwann cell activation. Sci Rep. 11:1840. DOI: 10.1038/s41598-021-81500-4. PMID: 33469141. PMCID: PMC7815837. PMID: 82fcadd287374c4b845bc72d2e93228b.32. Lin H, Huang H, Yu Y, Chen W, Zhang S, Zhang Y. 2021; Nerve growth factor regulates liver cancer cell polarity and motility. Mol Med Rep. 23:288. DOI: 10.3892/mmr.2021.11927. PMID: 33649819. PMCID: PMC7905331.33. Garrido MP, Torres I, Avila A, Chnaiderman J, Valenzuela-Valderrama M, Aramburo J, et al. 2020; NGF/TRKA decrease miR-145-5p levels in epithelial ovarian cancer cells. Int J Mol Sci. 21:7657. DOI: 10.3390/ijms21207657. PMID: 33081171. PMCID: PMC7589588. PMID: 4947c31d9dec4b11b37eb7ea92e29cf3.34. Faulkner S, Griffin N, Rowe CW, Jobling P, Lombard JM, Oliveira SM, et al. 2020; Nerve growth factor and its receptor tyrosine kinase TrkA are overexpressed in cervical squamous cell carcinoma. FASEB Bioadv. 2:398–408. DOI: 10.1096/fba.2020-00016. PMID: 32676580. PMCID: PMC7354692. PMID: c993e07b2b364f0ea4cb27aaf9d18bbb.35. Parimon T, Brauer R, Schlesinger SY, Xie T, Jiang D, Ge L, et al. 2018; Syndecan-1 controls lung tumorigenesis by regulating miRNAs packaged in exosomes. Am J Pathol. 188:1094–103. DOI: 10.1016/j.ajpath.2017.12.009. PMID: 29355516. PMCID: PMC5963476.36. Binder Gallimidi A, Nussbaum G, Hermano E, Weizman B, Meirovitz A, Vlodavsky I, et al. 2017; Syndecan-1 deficiency promotes tumor growth in a murine model of colitis-induced colon carcinoma. PLoS One. 12:e0174343. DOI: 10.1371/journal.pone.0174343. PMID: 28350804. PMCID: PMC5369774.37. Zhang Y, Wang Z, Liu J, Zhang S, Fei J, Li J, et al. 2017; Cell surface-anchored syndecan-1 ameliorates intestinal inflammation and neutrophil transmigration in ulcerative colitis. J Cell Mol Med. 21:13–25. Erratum in: J Cell Mol Med 2017; 21: 834. DOI: 10.1111/jcmm.12934. PMID: 27558380. PMCID: PMC5192823.38. Ibrahim SA, Gadalla R, El-Ghonaimy EA, Samir O, Mohamed HT, Hassan H, et al. 2017; Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol Cancer. 16:57. DOI: 10.1186/s12943-017-0621-z. PMID: 28270211. PMCID: PMC5341174.39. Yang Y, Tao X, Li CB, Wang CM. 2018; MicroRNA-494 acts as a tumor suppressor in pancreatic cancer, inhibiting epithelial-mesenchymal transition, migration and invasion by binding to SDC1. Int J Oncol. 53:1204–14. DOI: 10.3892/ijo.2018.4445. PMID: 29956739.40. Yu S, Lv H, Zhang H, Jiang Y, Hong Y, Xia R, et al. 2017; Heparanase-1-induced shedding of heparan sulfate from syndecan-1 in hepatocarcinoma cell facilitates lymphatic endothelial cell proliferation via VEGF-C/ERK pathway. Biochem Biophys Res Commun. 485:432–9. DOI: 10.1016/j.bbrc.2017.02.060. PMID: 28209511.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression and Clinical Significance of Heparanasein Osteosarcoma
- Application of Feedback Education to the Progression Notes Written by Medical Students in Surgical Clerkship
- Morphine as a suspect of aiding the propagation of cancer cells
- The study of nerve regeneration with infiltration of normal saline and nerve growth factor after vein graft to the resected sciatic nerve
- Effects of nerve cells and adhesion molecules on nerve conduit for peripheral nerve regeneration