Healthc Inform Res.
2022 Oct;28(4):387-393. 10.4258/hir.2022.28.4.387.
Development of a Secondary Dental-Specific Database for Active Learning of Genetics in Dentistry Programs
- Affiliations
-
- 1School of Dentistry, Faculty of Medicine and Dentistry, College of Health Sciences, University of Alberta, Edmonton, Canada
- KMID: 2535813
- DOI: http://doi.org/10.4258/hir.2022.28.4.387
Abstract
Objectives
Dental students study the genetics of tooth and facial development through didactic lectures only. Meanwhile, scientists’ knowledge of genetics is rapidly expanding, over and above what is commonly found in textbooks. Therefore, students studying dentistry are often unfamiliar with the burgeoning field of genetic data and biological databases. There is also a growing interest in applying active learning strategies to teach genetics in higher education. We developed a secondary database called “Genetics for Dentistry” to use as an active learning tool for teaching genetics in dentistry programs. The database archives genomic and proteomic data related to enamel and dentin formation.
Methods
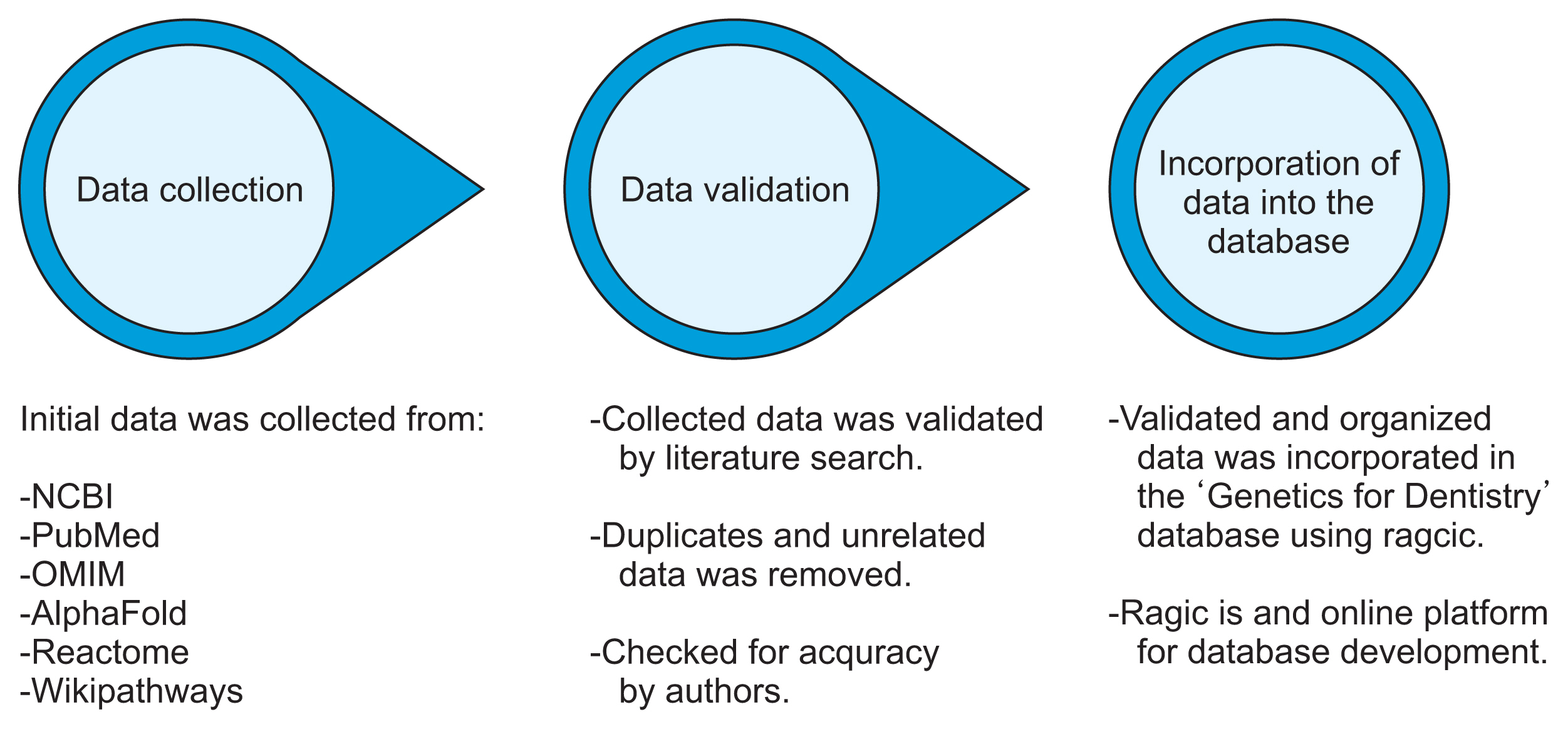
We took a systematic approach to identify, collect, and organize genomic and proteomic tooth development data from primary databases and literature searches. The data were checked for accuracy and exported to Ragic to create an interactive secondary database.
Results
“Genetics for Dentistry,” which is in its initial phase, contains information on all the human genes involved in enamel and dentin formation. Users can search the database by gene name, protein sequence, chromosomal location, and other keywords related to protein and gene function.
Conclusions
“Genetics for Dentistry” will be introduced as an active learning tool for teaching genetics at the School of Dentistry of the University of Alberta. Activities using the database will supplement lectures on genetics in the dentistry program. We hope that incorporating this database as an active learning tool will reduce students’ cognitive load in learning genetics and stimulate interest in new branches of science, including bioinformatics and precision dentistry.
Keyword
Figure
Reference
-
References
1. Thesleff I, Pirinen S. Dental anomalies: genetics. Encyclopedia of life sciences. Chichester, UK: John Wiley & Sons Ltd;2005.2. Klein ML, Nieminen P, Lammi L, Niebuhr E, Kreiborg S. Novel mutation of the initiation codon of PAX9 causes oligodontia. J Dent Res. 2005; 84(1):43–7. https://doi.org/10.1177/154405910508400107 .
Article3. Hooper JE, Feng W, Li H, Leach SM, Phang T, Siska C, et al. Systems biology of facial development: contributions of ectoderm and mesenchyme. Dev Biol. 2017; 426(1):97–114. https://doi.org/10.1016/j.ydbio.2017.03.025 .
Article4. Smith CE, Poulter JA, Antanaviciute A, Kirkham J, Brookes SJ, Inglehearn CF, et al. Amelogenesis imperfecta; genes, proteins, and pathways. Front Physiol. 2017; 8:435. https://doi.org/10.3389/fphys.2017.00435 .
Article5. Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014; 42(Database issue):D32–7. https://doi.org/10.1093/nar/gku1216 .
Article6. Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, et al. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002; 3(3):265–74. https://doi.org/10.1093/bib/3.3.265 .
Article7. Divaris K. Precision dentistry in early childhood: the central role of genomics. Dent Clin North Am. 2017; 61(3):619–25. https://doi.org/10.1016/j.cden.2017.02.008 .
Article8. Smith MK, Wood WB. Teaching genetics: past, present, and future. Genetics. 2016; 204(1):5–10. https://doi.org/10.1534/genetics.116.187138 .
Article9. Wood WB. Innovations in teaching undergraduate biology and why we need them. Annu Rev Cell Dev Biol. 2009; 25:93–112. https://doi.org/10.1146/annurev.cell-bio.24.110707.175306 .
Article10. Seymour E, Hewitt NM. Talking about leaving: why undergraduates leave the sciences. Boulder (CO): Westview Press;1996.11. Smith MK, Vinson EL, Smith JA, Lewin JD, Stetzer MR. A campus-wide study of STEM courses: new perspectives on teaching practices and perceptions. CBE Life Sci Educ. 2014; 13(4):624–35. https://doi.org/10.1187/cbe.14-06-0108 .
Article12. Lewin JD, Vinson EL, Stetzer MR, Smith MK. A campus-wide investigation of clicker implementation: the status of peer discussion in STEM classes. CBE Life Sci Educ. 2016. 15(1):ar6. https://doi.org/10.1187/cbe.15-10-0224 .
Article13. Hernandez-Serrano J, Choi I, Jonassen DH. Integrating constructivism and learning technologies. Spector JM, Anderson TM, editors. Integrated and holistic perspectives on learning, instruction and technology. Dordrecht, Netherlands: Springer;2000. 103–28. https://doi.org/10.1007/0-306-47584-7_7 .
Article14. Greening T. Building the constructivist toolbox: an exploration of cognitive technologies. Educ Technol. 1998; 38(2):23–35.15. Anderson LW, Krathwohl D. A taxonomy for learning, teaching, and assessing: a revision of Bloom’s taxonomy of educational objectives complete edition. New York (NY): Longman;2001.16. Jumper J, Hassabis D. Protein structure predictions to atomic accuracy with AlphaFold. Nat Methods. 2022; 19(1):11–2. https://doi.org/10.1038/s41592-021-01362-6 .
Article17. Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2016; 44(D1):D481–7. https://doi.org/10.1093/nar/gkv1351 .
Article18. Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008; 6(7):e184. https://doi.org/10.1371/journal.pbio.0060184 .
Article19. National Library of Medicine. National Center for Biotechnology Information [Internet]. Bethesda (MD): National Library of Medicine;c2022. [cited at 2022 Sep 30]. Available from: https://www.ncbi.nlm.nih.gov/ .20. Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005; 33(Database issue):D514–7. https://doi.org/10.1093/nar/gki033 .
Article21. NCBI Resource Coordinators. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016; 44(D1):D7–19. https://doi.org/10.1093/nar/gkv1290 .22. Ragic! Make everyone a data expert [Internet]. Seattle (WA): Ragic Inc;c2021. [cited at 2022 Sep 30]. Available from: https://www.ragic.com .23. Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, et al. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci U S A. 2014; 111(23):8410–5. https://doi.org/10.1073/pnas.1319030111 .
Article24. Prince M. Does active learning work? A review of the research. J Eng Educ. 2004; 93(3):223–31. https://doi.org/10.1002/j.2168-9830.2004.tb00809.x .
Article25. Lee WT, Jabot ME. Incorporating active learning techniques into a genetics class. J Coll Sci Teach. 2011; 40(4):94–100.26. Adams SM, Anderson KB, Coons JC, Smith RB, Meyer SM, Parker LS, et al. Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am J Pharm Educ. 2016; 80(1):3. https://doi.org/10.5688/ajpe8013 .
Article27. Neyhart JL, Watkins E. An active learning tool for quantitative genetics instruction using R and shiny. Nat Sci Educ. 2020; 49(1):e20026. https://doi.org/10.1002/nse2.20026 .
Article28. Carbone E. Students behaving badly in large classes. New Dir Teach Learn. 1999; (77):35–43.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Deep Learning in Dental Radiographic Imaging
- The Development of Problem-Based Learning Module for Clinical Dentistry in Dental Hygiene
- The Development of Case-Bank of Problem-Based Learning Program via the Internet for Medical Education
- Effect of repeated learning for two dental CAD software programs
- Development of Oral Pathology Learning Support System based on MMDB