Korean J Physiol Pharmacol.
2022 Nov;26(6):457-468. 10.4196/kjpp.2022.26.6.457.
Involvement of adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 in diallyl trisulfide-induced cytotoxicity in hepatocellular carcinoma cells
- Affiliations
-
- 1Department of Pathology, Renmin Hospital of Wuhan University, Wuhan 430060, China
- 2Department of Hepatobiliary & Laparascopic Surgery, Renmin Hospital of Wuhan University, Wuhan 430060, China
- 3Tongji Medical College Huazhong University of Science and Technology, School of Nursing, Wuhan 430030, China
- 4Department of Pathology and Pathophysiology, Wuhan University Taikang Medical School (School of Basic Medical Sciences), Wuhan 430071, China
- 5Department of Breast and Thyroid Surgery, Zhongnan Hospital of Wuhan University, Wuhan 430071, China
- KMID: 2534522
- DOI: http://doi.org/10.4196/kjpp.2022.26.6.457
Abstract
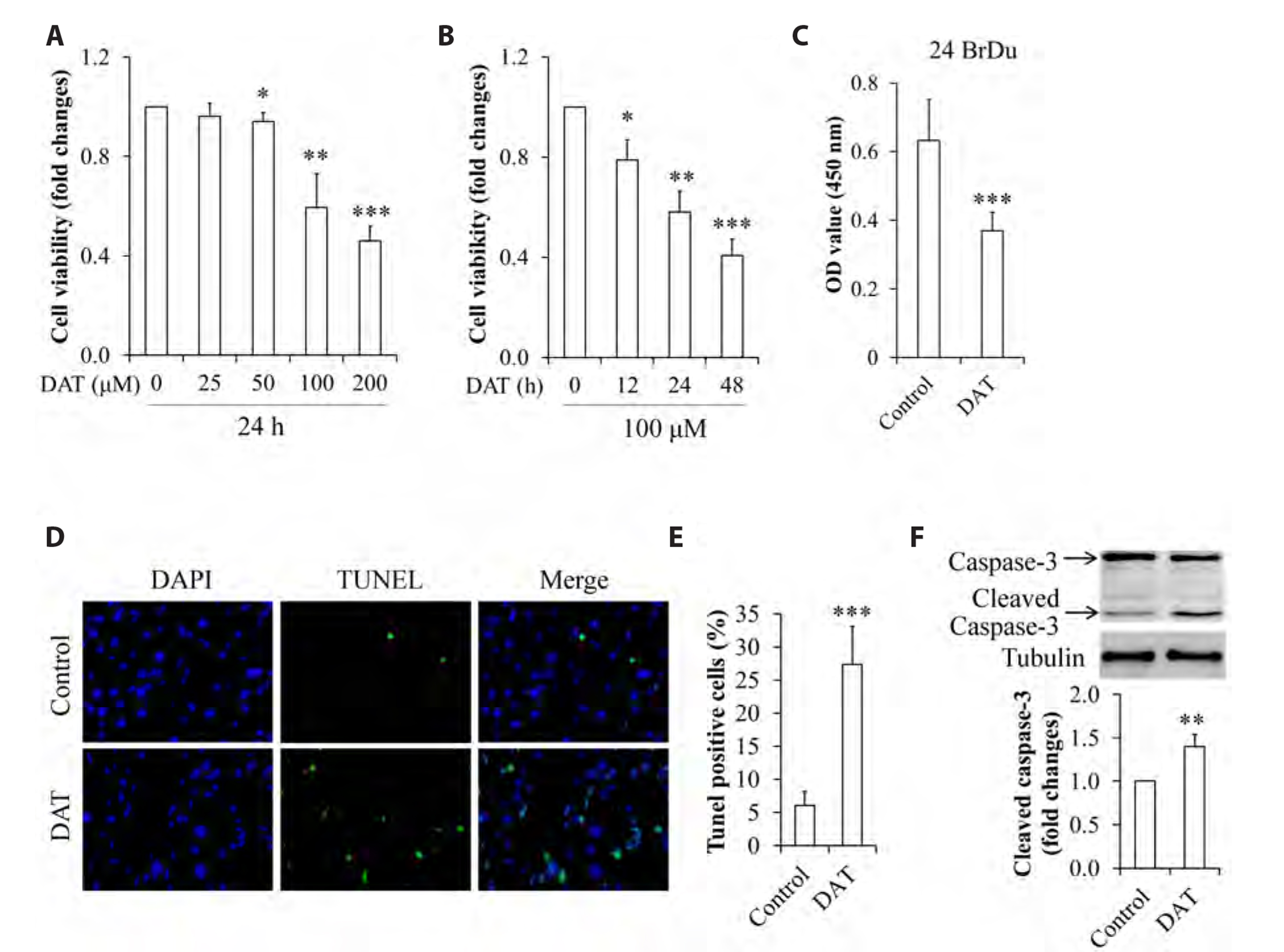
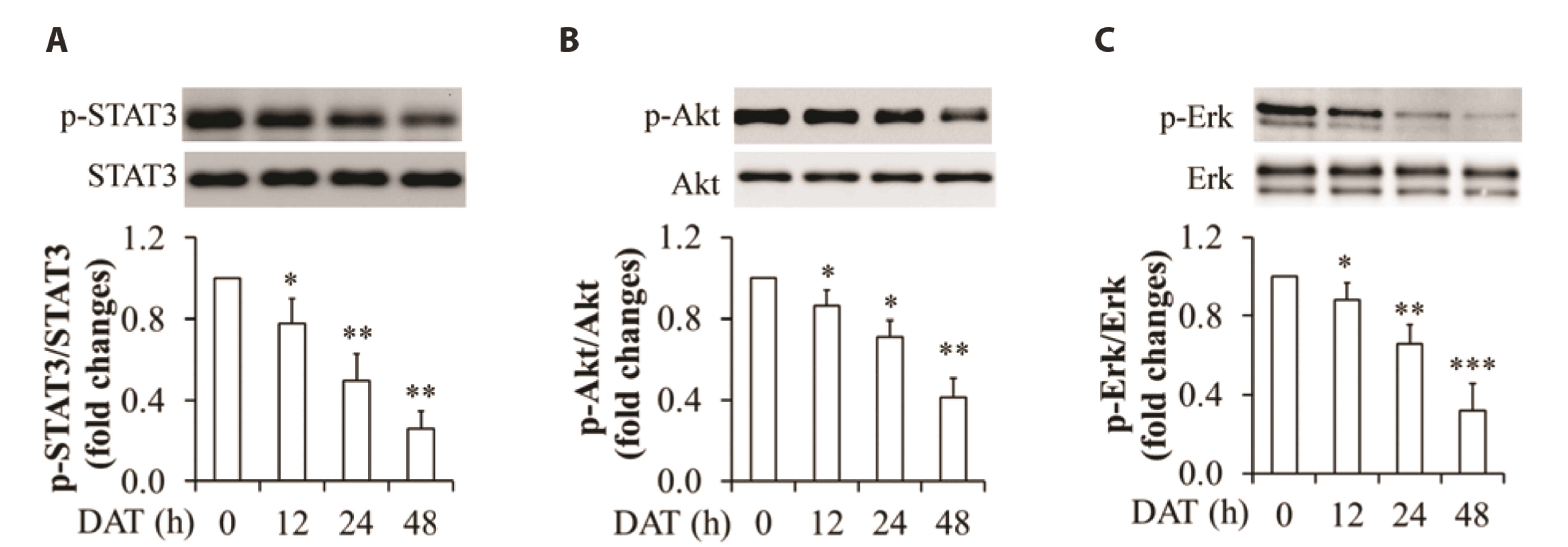
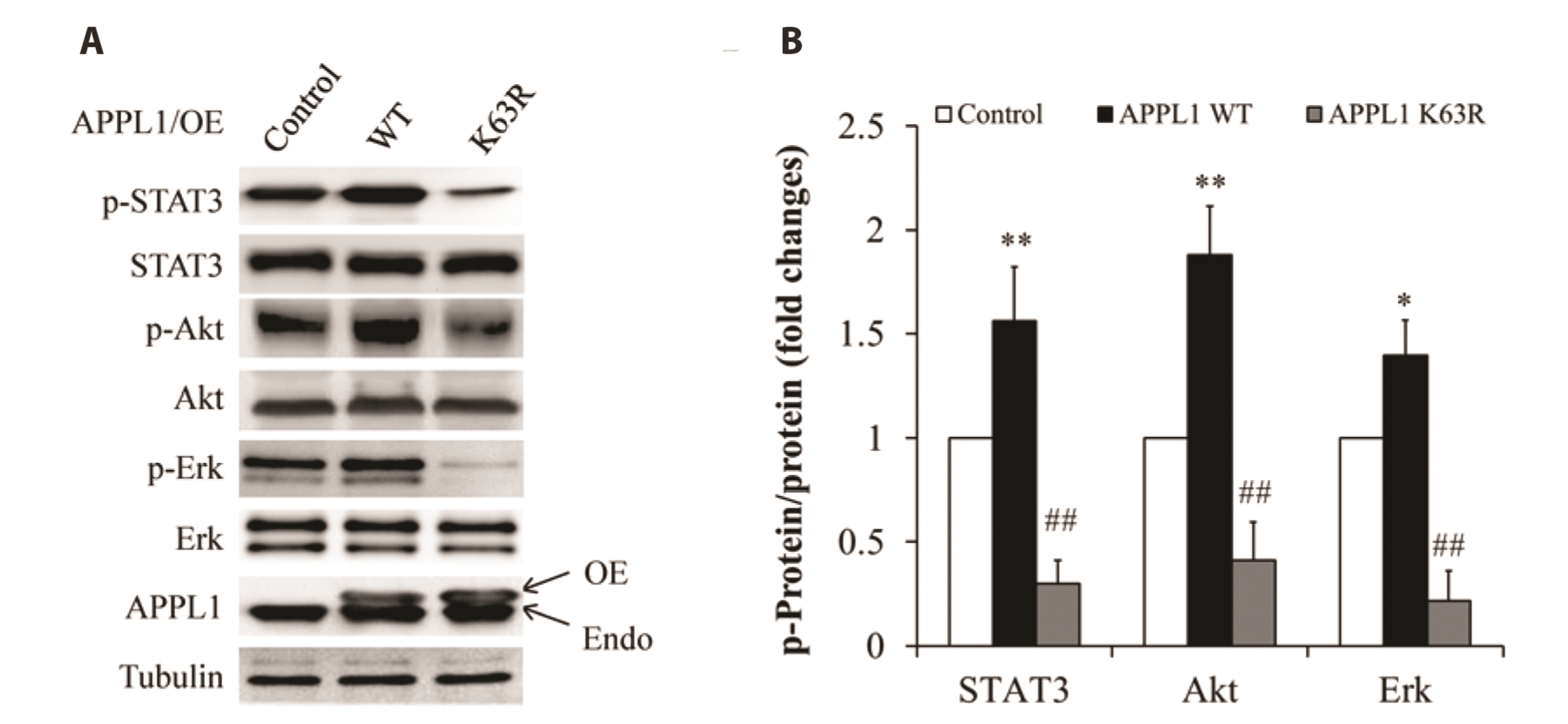
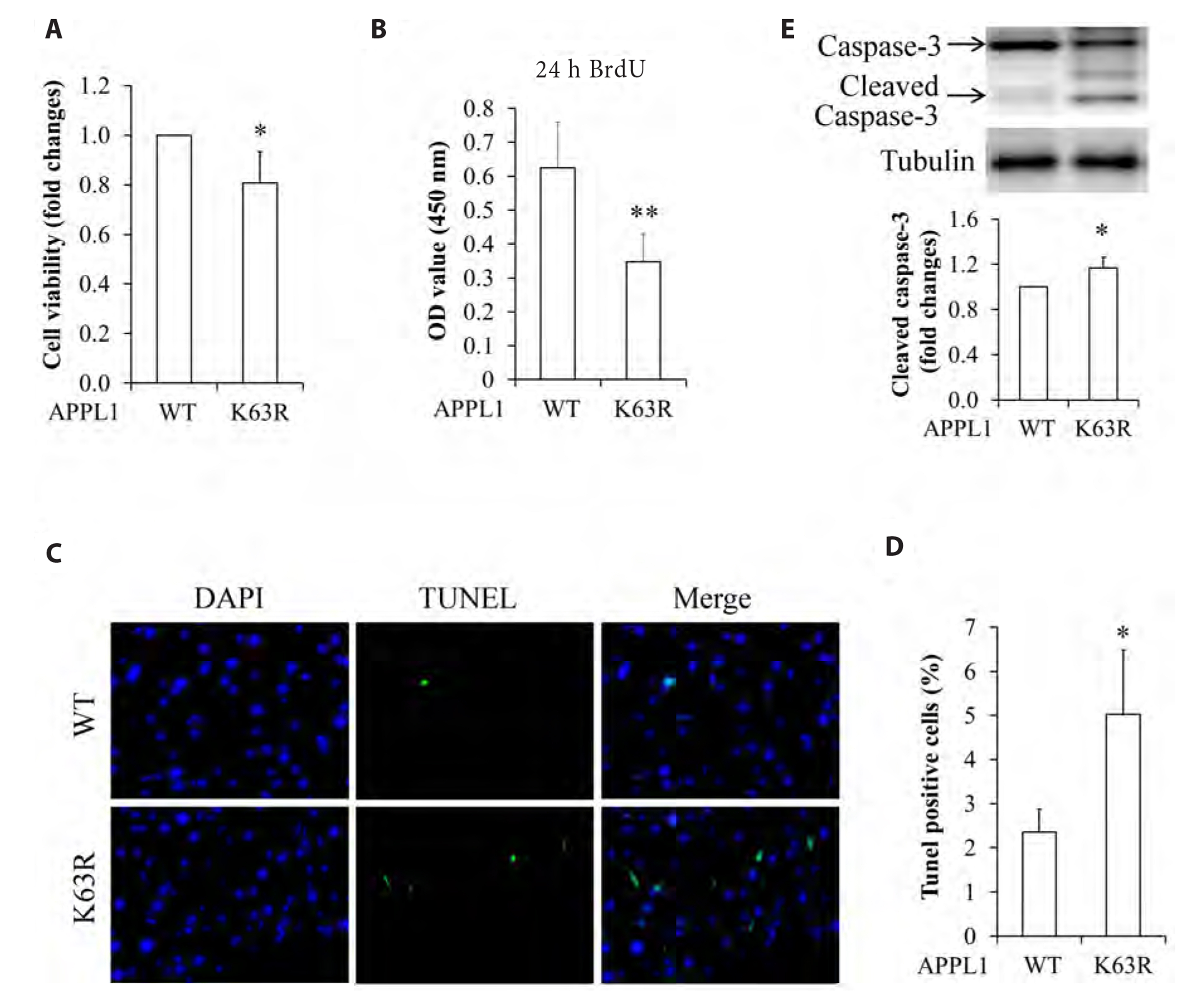
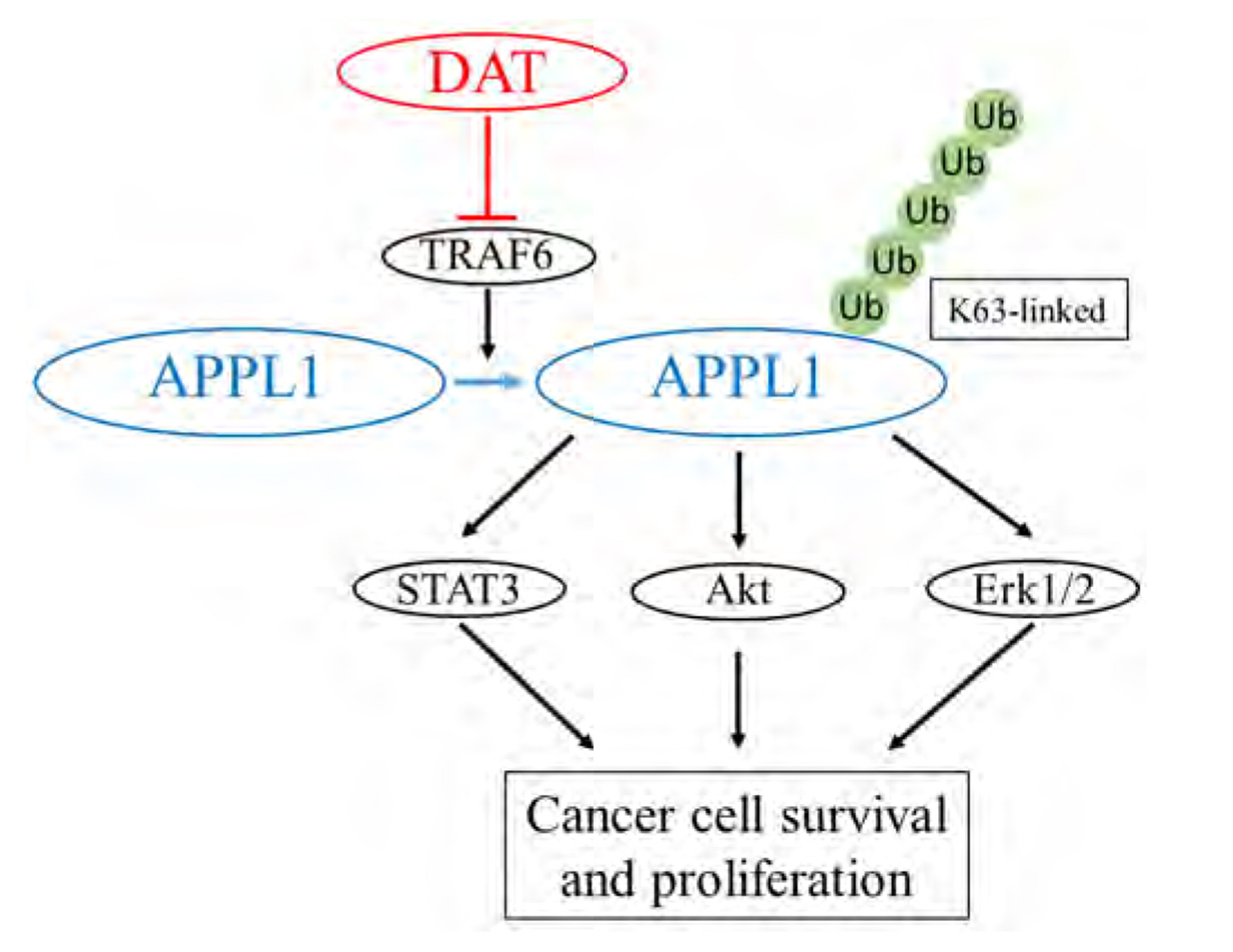
- It has been demonstrated that APPL1 (adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1) is involved in the regulation of several growth-related signaling pathways and thus closely associated with the development and progression of some cancers. Diallyl trisulfide (DAT), a garlic-derived bioactive compound, exerts selective cytotoxicity to various human cancer cells through interfering with pro-survival signaling pathways. However, whether and how DAT affects survival of human hepatocellular carcinoma (HCC) cells remain unclear. Herein, we tested the hypothesis of the involvement of APPL1 in DAT-induced cytotoxicity in HCC HepG2 cells. We found that Lys 63 (K63)-linked polyubiquitination of APPL1 was significantly decreased whereas phosphorylation of APPL1 at serine residues remained unchanged in DAT-treated HepG2 cells. Compared with wild-type APPL1, overexpression of APPL1 K63R mutant dramatically increased cell apoptosis and mitigated cell survival, along with a reduction of phosphorylation of STAT3, Akt, and Erk1/2. In addition, DAT administration markedly reduced protein levels of intracellular TNF receptor-associated factor 6 (TRAF6). Genetic inhibition of TRAF6 decreased K63-linked polyubiquitination of APPL1. Moreover, the cytotoxicity impacts of DAT on HepG2 cells were greatly attenuated by overexpression of wild-type APPL1. Taken together, these results suggest that APPL1 polyubiquitination probably mediates the inhibitory effects of DAT on survival of HepG2 cells by modulating STAT3, Akt, and Erk1/2 pathways.
Figure
Reference
-
1. Balogh J, Victor D 3rd, Asham EH, Burroughs SG, Boktour M, Saharia A, Li X, Ghobrial RM, Monsour HP Jr. 2016; Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 3:41–53. DOI: 10.2147/JHC.S61146. PMID: 27785449. PMCID: PMC5063561.
Article2. Chacko S, Samanta S. 2016; "Hepatocellular carcinoma: a life-threatening disease". Biomed Pharmacother. 84:1679–1688. DOI: 10.1016/j.biopha.2016.10.078. PMID: 27823920. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85001969730&origin=inward.
Article3. Best J, Schotten C, Theysohn JM, Wetter A, Müller S, Radünz S, Schulze M, Canbay A, Dechêne A, Gerken G. 2017; Novel implications in the treatment of hepatocellular carcinoma. Ann Gastroenterol. 30:23–32. DOI: 10.20524/aog.2016.0092. PMID: 28042235. PMCID: PMC5198244. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85008180773&origin=inward.4. Dutta R, Mahato RI. 2017; Recent advances in hepatocellular carcinoma therapy. Pharmacol Ther. 173:106–117. DOI: 10.1016/j.pharmthera.2017.02.010. PMID: 28174094. PMCID: PMC5777523. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85012930297&origin=inward.
Article5. Sapisochin G, Bruix J. 2017; Liver transplantation for hepatocellular carcinoma: outcomes and novel surgical approaches. Nat Rev Gastroenterol Hepatol. 14:203–217. DOI: 10.1038/nrgastro.2016.193. PMID: 28053342. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85008330472&origin=inward.
Article6. Adaki S, Adaki R, Shah K, Karagir A. 2014; Garlic: review of literature. Indian J Cancer. 51:577–581. DOI: 10.4103/0019-509X.175383. PMID: 26842201. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84958178966&origin=inward.
Article7. Yu L, Li S, Tang X, Li Z, Zhang J, Xue X, Han J, Liu Y, Zhang Y, Zhang Y, Xu Y, Yang Y, Wang H. 2017; Diallyl trisulfide ameliorates myocardial ischemia-reperfusion injury by reducing oxidative stress and endoplasmic reticulum stress-mediated apoptosis in type 1 diabetic rats: role of SIRT1 activation. Apoptosis. 22:942–954. DOI: 10.1007/s10495-017-1378-y. PMID: 28455824. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85018294902&origin=inward.
Article8. Zhu X, Zhang F, Zhou L, Kong D, Chen L, Lu Y, Zheng S. 2014; Diallyl trisulfide attenuates carbon tetrachloride-caused liver injury and fibrogenesis and reduces hepatic oxidative stress in rats. Naunyn Schmiedebergs Arch Pharmacol. 387:445–455. DOI: 10.1007/s00210-014-0959-3. PMID: 24557053. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84899965294&origin=inward.
Article9. Tsai CY, Wen SY, Shibu MA, Yang YC, Peng H, Wang B, Wei YM, Chang HY, Lee CY, Huang CY, Kuo WW. 2015; Diallyl trisulfide protects against high glucose-induced cardiac apoptosis by stimulating the production of cystathionine gamma-lyase-derived hydrogen sulfide. Int J Cardiol. 195:300–310. DOI: 10.1016/j.ijcard.2015.05.111. PMID: 26056963. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84937611169&origin=inward.
Article10. Hayashida R, Kondo K, Morita S, Unno K, Shintani S, Shimizu Y, Calvert JW, Shibata R, Murohara T. 2017; Diallyl trisulfide augments ischemia-induced angiogenesis via an endothelial nitric oxide synthase-dependent mechanism. Circ J. 81:870–878. DOI: 10.1253/circj.CJ-16-1097. PMID: 28216514. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85019773175&origin=inward.
Article11. Chen LY, Chen Q, Cheng YF, Jin HH, Kong DS, Zhang F, Wu L, Shao JJ, Zheng SZ. 2016; Diallyl trisulfide attenuates ethanol-induced hepatic steatosis by inhibiting oxidative stress and apoptosis. Biomed Pharmacother. 79:35–43. DOI: 10.1016/j.biopha.2016.01.009. PMID: 27044810. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84961894562&origin=inward.
Article12. Wang S, Li M, Wang X, Li X, Yin H, Jiang L, Han W, Irving G, Zeng T, Xie K. 2017; Diallyl trisulfide attenuated n-hexane induced neurotoxicity in rats by modulating P450 enzymes. Chem Biol Interact. 265:1–7. DOI: 10.1016/j.cbi.2017.01.013. PMID: 28115069. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85010399976&origin=inward.
Article13. Lii CK, Huang CY, Chen HW, Chow MY, Lin YR, Huang CS, Tsai CW. 2012; Diallyl trisulfide suppresses the adipogenesis of 3T3-L1 preadipocytes through ERK activation. Food Chem Toxicol. 50:478–484. DOI: 10.1016/j.fct.2011.11.020. PMID: 22137902. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84863298355&origin=inward.
Article14. Zhang F, Zhang Y, Wang K, Liu G, Yang M, Zhao Z, Li S, Cai J, Cao J. 2016; Protective effect of diallyl trisulfide against naphthalene-induced oxidative stress and inflammatory damage in mice. Int J Immunopathol Pharmacol. 29:205–216. DOI: 10.1177/0394632015627160. PMID: 26813860. PMCID: PMC5806724. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84966701534&origin=inward.
Article15. Kiesel VA, Stan SD. 2017; Diallyl trisulfide, a chemopreventive agent from Allium vegetables, inhibits alpha-secretases in breast cancer cells. Biochem Biophys Res Commun. 484:833–838. DOI: 10.1016/j.bbrc.2017.01.184. PMID: 28161636. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85011878196&origin=inward.
Article16. Jiang X, Zhu X, Liu N, Xu H, Zhao Z, Li S, Li S, Cai J, Cao J. 2017; Diallyl trisulfide inhibits growth of NCI-H460 in vitro and in vivo, and ameliorates cisplatin-induced oxidative injury in the treatment of lung carcinoma in xenograft mice. Int J Biol Sci. 13:167–178. DOI: 10.7150/ijbs.16828. PMID: 28255269. PMCID: PMC5332871. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85011831927&origin=inward.
Article17. Jiang XY, Zhu XS, Xu HY, Zhao ZX, Li SY, Li SZ, Cai JH, Cao JM. 2017; Diallyl trisulfide suppresses tumor growth through the attenuation of Nrf2/Akt and activation of p38/JNK and potentiates cisplatin efficacy in gastric cancer treatment. Acta Pharmacol Sin. 38:1048–1058. DOI: 10.1038/aps.2016.176. PMID: 28344324. PMCID: PMC5519247. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85021820280&origin=inward.
Article18. Hosono T, Fukao T, Ogihara J, Ito Y, Shiba H, Seki T, Ariga T. 2005; Diallyl trisulfide suppresses the proliferation and induces apoptosis of human colon cancer cells through oxidative modification of beta-tubulin. J Biol Chem. 280:41487–41493. DOI: 10.1074/jbc.M507127200. PMID: 16219763. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=29244484434&origin=inward.
Article19. Lai KC, Hsu SC, Yang JS, Yu CC, Lein JC, Chung JG. 2015; Diallyl trisulfide inhibits migration, invasion and angiogenesis of human colon cancer HT-29 cells and umbilical vein endothelial cells, and suppresses murine xenograft tumour growth. J Cell Mol Med. 19:474–484. DOI: 10.1111/jcmm.12486. PMID: 25403643. PMCID: PMC4407594. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84921786219&origin=inward.
Article20. Xiao D, Choi S, Johnson DE, Vogel VG, Johnson CS, Trump DL, Lee YJ, Singh SV. 2004; Diallyl trisulfide-induced apoptosis in human prostate cancer cells involves c-Jun N-terminal kinase and extracellular-signal regulated kinase-mediated phosphorylation of Bcl-2. Oncogene. 23:5594–5606. DOI: 10.1038/sj.onc.1207747. PMID: 15184882. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=3342895063&origin=inward.
Article21. Xiao D, Lew KL, Kim YA, Zeng Y, Hahm ER, Dhir R, Singh SV. 2006; Diallyl trisulfide suppresses growth of PC-3 human prostate cancer xenograft in vivo in association with Bax and Bak induction. Clin Cancer Res. 12:6836–6843. DOI: 10.1158/1078-0432.CCR-06-1273. PMID: 17121905. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33845299153&origin=inward.
Article22. Li Y, Zhang J, Zhang L, Si M, Yin H, Li J. 2013; Diallyl trisulfide inhibits proliferation, invasion and angiogenesis of osteosarcoma cells by switching on suppressor microRNAs and inactivating of Notch-1 signaling. Carcinogenesis. 34:1601–1610. DOI: 10.1093/carcin/bgt065. PMID: 23430952. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84880320805&origin=inward.
Article23. Zhang ZM, Yang XY, Deng SH, Xu W, Gao HQ. 2007; Anti-tumor effects of polybutylcyanoacrylate nanoparticles of diallyl trisulfide on orthotopic transplantation tumor model of hepatocellular carcinoma in BALB/c nude mice. Chin Med J (Engl). 120:1336–1342. DOI: 10.1097/00029330-200708010-00008. PMID: 17711740. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34548296990&origin=inward.
Article24. Deepa SS, Dong LQ. 2009; APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab. 296:E22–E36. DOI: 10.1152/ajpendo.90731.2008. PMID: 18854421. PMCID: PMC2636986. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=58249105213&origin=inward.
Article25. Liu Z, Xiao T, Peng X, Li G, Hu F. 2017; APPLs: more than just adiponectin receptor binding proteins. Cell Signal. 32:76–84. DOI: 10.1016/j.cellsig.2017.01.018. PMID: 28108259. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85010297595&origin=inward.
Article26. Johnson IR, Parkinson-Lawrence EJ, Keegan H, Spillane CD, Barry-O'Crowley J, Watson WR, Selemidis S, Butler LM, O'Leary JJ, Brooks DA. 2015; Endosomal gene expression: a new indicator for prostate cancer patient prognosis? Oncotarget. 6:37919–37929. DOI: 10.18632/oncotarget.6114. PMID: 26473288. PMCID: PMC4741974. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84947786217&origin=inward.
Article27. Song J, Mu Y, Li C, Bergh A, Miaczynska M, Heldin CH, Landström M. 2016; APPL proteins promote TGFβ-induced nuclear transport of the TGFβ type I receptor intracellular domain. Oncotarget. 7:279–292. DOI: 10.18632/oncotarget.6346. PMID: 26583432. PMCID: PMC4807998. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85020883234&origin=inward.
Article28. Ding Y, Cao Y, Wang B, Wang L, Zhang Y, Zhang D, Chen X, Li M, Wang C. 2016; APPL1-mediating leptin signaling contributes to proliferation and migration of cancer cells. PLoS One. 11:e0166172. DOI: 10.1371/journal.pone.0166172. PMID: 27820851. PMCID: PMC5098739. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84994589804&origin=inward.
Article29. Hennig J, McShane MP, Cordes N, Eke I. 2014; APPL proteins modulate DNA repair and radiation survival of pancreatic carcinoma cells by regulating ATM. Cell Death Dis. 5:e1199. DOI: 10.1038/cddis.2014.167. PMID: 24763056. PMCID: PMC4001316. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84901045038&origin=inward.
Article30. Kunkel TA, Roberts JD, Zakour RA. 1987; Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367–382. DOI: 10.1016/0076-6879(87)54085-X. PMID: 3323813. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0023613953&origin=inward.
Article31. Cheng KK, Lam KS, Wang Y, Wu D, Zhang M, Wang B, Li X, Hoo RL, Huang Z, Sweeney G, Xu A. 2013; TRAF6-mediated ubiquitination of APPL1 enhances hepatic actions of insulin by promoting the membrane translocation of Akt. Biochem J. 455:207–216. DOI: 10.1042/BJ20130760. PMID: 23909487. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84884787995&origin=inward.
Article32. Wang C, Xin X, Xiang R, Ramos FJ, Liu M, Lee HJ, Chen H, Mao X, Kikani CK, Liu F, Dong LQ. 2009; Yin-Yang regulation of adiponectin signaling by APPL isoforms in muscle cells. J Biol Chem. 284:31608–31615. DOI: 10.1074/jbc.M109.010355. PMID: 19661063. PMCID: PMC2797231. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=70450265225&origin=inward.
Article33. Ding Y, Zhang D, Wang B, Zhang Y, Wang L, Chen X, Li M, Tang Z, Wang C. 2016; APPL1-mediated activation of STAT3 contributes to inhibitory effect of adiponectin on hepatic gluconeogenesis. Mol Cell Endocrinol. 433:12–19. DOI: 10.1016/j.mce.2016.05.021. PMID: 27246173. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84973326543&origin=inward.
Article34. Guan F, Ding Y, Zhang Y, Zhou Y, Li M, Wang C. 2016; Curcumin suppresses proliferation and migration of MDA-MB-231 breast cancer cells through autophagy-dependent Akt degradation. PLoS One. 11:e0146553. DOI: 10.1371/journal.pone.0146553. PMID: 26752181. PMCID: PMC4708990. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84954567599&origin=inward.
Article35. Ding Y, Wang B, Chen X, Zhou Y, Ge J. 2017; Staurosporine suppresses survival of HepG2 cancer cells through Omi/HtrA2-mediated inhibition of PI3K/Akt signaling pathway. Tumour Biol. 39:1010428317694317. DOI: 10.1177/1010428317694317. PMID: 28349827. PMID: 26a7cdaef35d4ddb81d28370ec20d244. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=85016925370&origin=inward.
Article36. Bai J, Cederbaum AI. 2006; Cycloheximide protects HepG2 cells from serum withdrawal-induced apoptosis by decreasing p53 and phosphorylated p53 levels. J Pharmacol Exp Ther. 319:1435–1443. DOI: 10.1124/jpet.106.110007. PMID: 16971506. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33751188946&origin=inward.37. Pilecka I, Sadowski L, Kalaidzidis Y, Miaczynska M. 2011; Recruitment of APPL1 to ubiquitin-rich aggresomes in response to proteasomal impairment. Exp Cell Res. 317:1093–1107. DOI: 10.1016/j.yexcr.2011.02.002. PMID: 21320486. PMCID: PMC3072527. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=79953069853&origin=inward.
Article38. Xiao D, Singh SV. 2006; Diallyl trisulfide, a constituent of processed garlic, inactivates Akt to trigger mitochondrial translocation of BAD and caspase-mediated apoptosis in human prostate cancer cells. Carcinogenesis. 27:533–540. DOI: 10.1093/carcin/bgi228. PMID: 16169930. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=33644869135&origin=inward.
Article39. Shrotriya S, Kundu JK, Na HK, Surh YJ. 2010; Diallyl trisulfide inhibits phorbol ester-induced tumor promotion, activation of AP-1, and expression of COX-2 in mouse skin by blocking JNK and Akt signaling. Cancer Res. 70:1932–1940. Erratum in: Cancer Res. 2010;70:3414. DOI: 10.1158/0008-5472.CAN-09-3501. PMID: 20179211. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77950280661&origin=inward.
Article40. Chandra-Kuntal K, Singh SV. 2010; Diallyl trisulfide inhibits activation of signal transducer and activator of transcription 3 in prostate cancer cells in culture and in vivo. Cancer Prev Res (Phila). 3:1473–1483. DOI: 10.1158/1940-6207.CAPR-10-0123. PMID: 20959517. PMCID: PMC2988081. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=78649238180&origin=inward.
Article41. Wang H, Sun N, Li X, Li K, Tian J, Li J. 2016; Diallyl trisulfide induces osteosarcoma cell apoptosis through reactive oxygen species-mediated downregulation of the PI3K/Akt pathway. Oncol Rep. 35:3648–3658. DOI: 10.3892/or.2016.4722. PMID: 27035545. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84964910272&origin=inward.
Article42. Saxena NK, Sharma D, Ding X, Lin S, Marra F, Merlin D, Anania FA. 2007; Concomitant activation of the JAK/STAT, PI3K/AKT, and ERK signaling is involved in leptin-mediated promotion of invasion and migration of hepatocellular carcinoma cells. Cancer Res. 67:2497–2507. DOI: 10.1158/0008-5472.CAN-06-3075. PMID: 17363567. PMCID: PMC2925446. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=34047250349&origin=inward.
Article43. Gant-Branum RL, Broussard JA, Mahsut A, Webb DJ, McLean JA. 2010; Identification of phosphorylation sites within the signaling adaptor APPL1 by mass spectrometry. J Proteome Res. 9:1541–1548. DOI: 10.1021/pr901043e. PMID: 20095645. PMCID: PMC2845304. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=77949780036&origin=inward.
Article44. Holmes RM, Yi Z, De Filippis E, Berria R, Shahani S, Sathyanarayana P, Sherman V, Fujiwara K, Meyer C, Christ-Roberts C, Hwang H, Finlayson J, Dong LQ, Mandarino LJ, Bajaj M. 2011; Increased abundance of the adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain and leucine zipper motif (APPL1) in patients with obesity and type 2 diabetes: evidence for altered adiponectin signalling. Diabetologia. 54:2122–2131. DOI: 10.1007/s00125-011-2173-x. PMID: 21562756. PMCID: PMC3131511. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=79960912411&origin=inward.
Article45. Liu M, Zhou L, Wei L, Villarreal R, Yang X, Hu D, Riojas RA, Holmes BM, Langlais PR, Lee H, Dong LQ. 2012; Phosphorylation of adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1) at Ser430 mediates endoplasmic reticulum (ER) stress-induced insulin resistance in hepatocytes. J Biol Chem. 287:26087–26093. DOI: 10.1074/jbc.M112.372292. PMID: 22685300. PMCID: PMC3406692. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84864390268&origin=inward.
Article46. Wang J, Lu W, Chen L, Zhang P, Qian T, Cao W, Luo J. 2016; Serine 707 of APPL1 is critical for the synaptic NMDA receptor-mediated Akt phosphorylation signaling pathway. Neurosci Bull. 32:323–330. DOI: 10.1007/s12264-016-0042-9. PMID: 27300007. PMCID: PMC5563782. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84974851680&origin=inward.
Article47. Komander D. 2009; The emerging complexity of protein ubiquitination. Biochem Soc Trans. 37(Pt 5):937–953. DOI: 10.1042/BST0370937. PMID: 19754430. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=70350150000&origin=inward.
Article48. Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. 2009; Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145. DOI: 10.1016/j.cell.2009.01.041. PMID: 19345192. PMCID: PMC2668214. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=63049125531&origin=inward.
Article49. Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000; Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 103:351–361. DOI: 10.1016/S0092-8674(00)00126-4. PMID: 11057907. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0034644474&origin=inward.
Article50. Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002; RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141. DOI: 10.1038/nature00991. PMID: 12226657. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=0037068455&origin=inward.
Article51. Lauwers E, Jacob C, André B. 2009; K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol. 185:493–502. DOI: 10.1083/jcb.200810114. PMID: 19398763. PMCID: PMC2700384. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=65649128660&origin=inward.
Article52. Silva GM, Finley D, Vogel C. 2015; K63 polyubiquitination is a new modulator of the oxidative stress response. Nat Struct Mol Biol. 22:116–123. DOI: 10.1038/nsmb.2955. PMID: 25622294. PMCID: PMC4318705. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84926417515&origin=inward.
Article53. Zu Y, Yang Y, Zhu J, Bo X, Hou S, Zhang B, Qiu J, Zheng J. 2016; MiR-146a suppresses hepatocellular carcinoma by downregulating TRAF6. Am J Cancer Res. 6:2502–2513. PMID: 27904767. PMCID: PMC5126269.54. Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu YR, Yang J, Ding H, Qin GH, Yang LH, Dang YW, Yang H, Chen G. 2016; Relationship between TRAF6 and deterioration of HCC: an immunohistochemical and in vitro study. Cancer Cell Int. 16:76. Erratum in: Cancer Cell Int. 2020;20:60. DOI: 10.1186/s12935-016-0352-z. PMID: 27708550. PMCID: PMC5041287. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84988930452&origin=inward.
Article55. Shigemi Z, Furukawa Y, Hosokawa K, Minami S, Matsuhiro J, Nakata S, Watanabe T, Kagawa H, Nakagawa K, Takeda H, Fujimuro M. 2016; Diallyl trisulfide induces apoptosis by suppressing NF-κB signaling through destabilization of TRAF6 in primary effusion lymphoma. Int J Oncol. 48:293–304. DOI: 10.3892/ijo.2015.3247. PMID: 26647777. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84953329084&origin=inward.
Article56. Schmitz KJ, Wohlschlaeger J, Lang H, Sotiropoulos GC, Malago M, Steveling K, Reis H, Cicinnati VR, Schmid KW, Baba HA. 2008; Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol. 48:83–90. DOI: 10.1016/j.jhep.2007.08.018. PMID: 17998146. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=36549013178&origin=inward.
Article57. Yang J, Cai X, Lu W, Hu C, Xu X, Yu Q, Cao P. 2013; Evodiamine inhibits STAT3 signaling by inducing phosphatase shatterproof 1 in hepatocellular carcinoma cells. Cancer Lett. 328:243–251. DOI: 10.1016/j.canlet.2012.09.019. PMID: 23032719. PMID: https://www.scopus.com/inward/record.uri?partnerID=HzOxMe3b&scp=84870364791&origin=inward.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The centrosomal localization of KM-HN-1 (MGC33607) depends on the leucine zipper motif and the C-terminal coiled-coil domain
- Association of Killer Cell Ig-like Receptor (KIR) with an Adaptor Protein Shc
- Exploring the role and mechanisms of diallyl trisulfide and diallyl disulfide in chronic constriction-induced neuropathic pain in rats
- Ubiquitylation of Fe65 adaptor protein by neuronal precursor cell expressed developmentally down regulated 4-2 (Nedd4-2) via the WW domain interaction with Fe65
- Gene Expression Changes by Diallyl Trisulfide Administration in Chemically-induced Mammary Tumors in Rats