Healthc Inform Res.
2022 Jul;28(3):276-283. 10.4258/hir.2022.28.3.276.
Text Mining of Biomedical Articles Using the Konstanz Information Miner (KNIME) Platform: Hemolytic Uremic Syndrome as a Case Study
- Affiliations
-
- 1Facultad de Medicina, Instituto de Fisiología y Biofísica Bernardo Houssay (IFIBIO Houssay), CONICET–Universidad de Buenos Aires, Buenos Aires, Argentina
- KMID: 2532455
- DOI: http://doi.org/10.4258/hir.2022.28.3.276
Abstract
Objectives
Automated systems for information extraction are becoming very useful due to the enormous scale of the existing literature and the increasing number of scientific articles published worldwide in the field of medicine. We aimed to develop an accessible method using the open-source platform KNIME to perform text mining (TM) on indexed publications. Material from scientific publications in the field of life sciences was obtained and integrated by mining information on hemolytic uremic syndrome (HUS) as a case study.
Methods
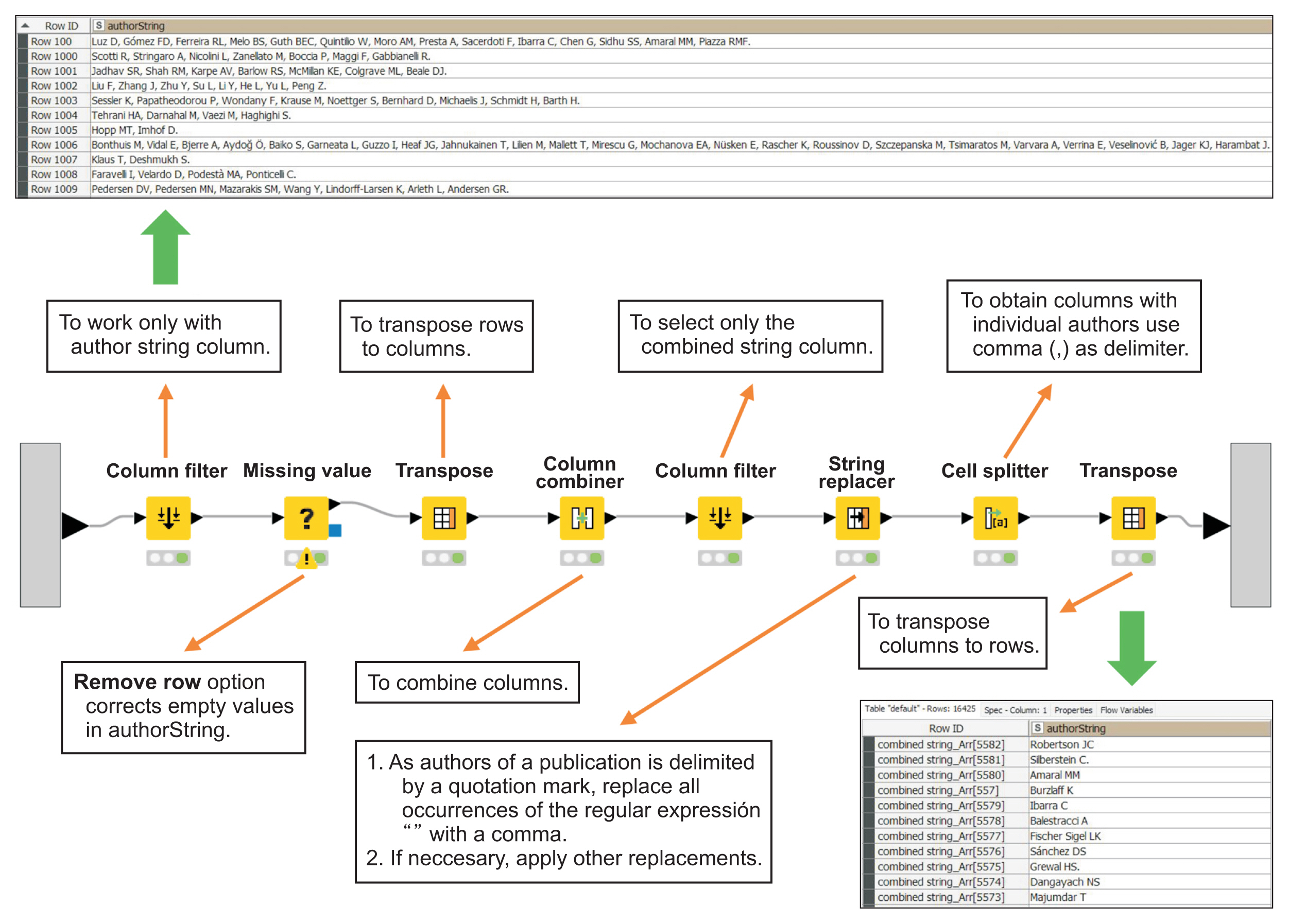
Text retrieved from Europe PubMed Central (PMC) was processed using specific KNIME nodes. The results were presented in the form of tables or graphical representations. Data could also be compared with those from other sources.
Results
By applying TM to the scientific literature on HUS as a case study, and by selecting various fields from scientific articles, it was possible to obtain a list of individual authors of publications, build bags of words and study their frequency and temporal use, discriminate topics (HUS vs. atypical HUS) in an unsupervised manner, and cross-reference information with a list of FDA-approved drugs.
Conclusions
Following the instructions in the tutorial, researchers without programming skills can successfully perform TM on the indexed scientific literature. This methodology, using KNIME, could become a useful tool for performing statistics, analyzing behaviors, following trends, and making forecast related to medical issues. The advantages of TM using KNIME include enabling the integration of scientific information, helping to carry out reviews, and optimizing the management of resources dedicated to basic and clinical research.
Keyword
Figure
Reference
-
References
1. Renganathan V. Text mining in biomedical domain with emphasis on document clustering. Healthc Inform Res. 2017; 23(3):141–6. https://doi.org/10.4258/hir.2017.23.3.141.
Article2. Tshitoyan V, Dagdelen J, Weston L, Dunn A, Rong Z, Kononova O, et al. Unsupervised word embeddings capture latent knowledge from materials science literature. Nature. 2019; 571(7763):95–8. https://doi.org/10.1038/s41586-019-1335-8.
Article3. Viceconti M, Hunter P. The virtual physiological human: ten years after. Annu Rev Biomed Eng. 2016; 18:103–23. https://doi.org/10.1146/annurev-bioeng-110915-114742.
Article4. Hotho A, Nurnberger A, Paaß G. A brief survey of text mining. LDV Forum. 2005; 20(1):19–62.5. Dorr RA, Casal JJ, Toriano R. Minería de texto en publicaciones científicas con autores argentinos [Text mining in scientific publications with Argentine authors]. Medicina (B Aires). 2021; 81(2):214–23.6. Dorr RA, Silberstein C, Ibarra C, Toriano R. Obtaining new information on hemolytic uremic syndrome by text mining. Medicina (B Aires). 2022; 82(4):513–24. PMID: 35904906.7. Jokiranta TS. HUS and atypical HUS. Blood. 2017; 129(21):2847–56. https://doi.org/10.1182/blood-2016-11-709865.
Article8. Exeni RA, Fernandez-Brando RJ, Santiago AP, Fiorentino GA, Exeni AM, Ramos MV, et al. Pathogenic role of inflammatory response during Shiga toxin-associated hemolytic uremic syndrome (HUS). Pediatr Nephrol. 2018; 33(11):2057–71. https://doi.org/10.1007/s00467-017-3876-0.
Article9. Newman D, Asuncion A, Smyth P, Welling M. Distributed algorithms for topic models. J Mach Learn Res. 2009; 10:1801–28.10. Yao L, Mimno D, McCallum A. Efficient methods for topic model inference on streaming document collections. In : Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2009 Jun 28–Jul 1; Paris, France. p. 937–46. https://doi.org/10.1145/1557019.1557121.
Article11. Qundus JA, Peikert S, Paschke A. AI supported topic modeling using KNIME-workflows [Internet]. Ithaca (NY): arXiv.org;2021. [cited at 2022 May 2]. Available from: https://arxiv.org/abs/2104.09428.12. Patel S, Patel A, Patel M, Shah U, Patel M, Solanki N, et al. Review and analysis of massively registered clinical trials of COVID-19 using the text mining approach. Rev Recent Clin Trials. 2021; 16(3):242–57. https://doi.org/10.2174/1574887115666201202110919.
Article13. Ordenes FV, Silipo R. Machine learning for marketing on the KNIME Hub: the development of a live repository for marketing applications. J Bus Res. 2021; 137:393–410. https://doi.org/10.1016/j.jbusres.2021.08.036.
Article14. Feltrin L. KNIME an open source solution for predictive analytics in the geosciences [software and data sets]. IEEE Geosci Remote Sens Mag. 2015; 3(4):28–38. https://doi.org/10.1109/MGRS.2015.2496160.
Article15. Vijayan R. Teaching and learning during the COVID-19 pandemic: a topic modeling study. Educ Sci. 2021; 11(7):347. https://doi.org/10.3390/educsci11070347.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Text Mining in Biomedical Domain with Emphasis on Document Clustering
- Codeless Deep Learning of COVID-19 Chest X-Ray Image Dataset with KNIME Analytics Platform
- A clinical aspect of the hemolytic uremic syndrome
- A Case of the Diarrhea-associated Hemolytic Uremic Syndrome Developing Simultaneously with an Acute Appendicitis
- A Case of Microangiopathic Hemolytic Anemia after Myxoma Excision and Mitral Valve Repair Presenting as Hemolytic Uremic Syndrome