Lab Med Online.
2021 Jan;11(1):25-31. 10.47429/lmo.2021.11.1.25.
Status of Next-Generation Sequencing-Based Genetic Diagnosis in Hematologic Malignancies in Korea (2017-2018)
- Affiliations
-
- 1Department of Laboratory Medicine, Yonsei University School of Medicine, Seoul, Korea
- 2Department of Laboratory Medicine, Inje University College of Medicine, Busan, Korea
- 3Department of Laboratory Medicine, Ewha Womans University College of Medicine, Seoul, Korea
- 4Department of Laboratory Medicine, Korea University School of Medicine, Seoul, Korea
- 5Department of Laboratory Medicine, College of Medicine, The Catholic University of Korea, Seoul, Korea
- 6Department of Laboratory Medicine, University of Ulsan College of Medicine, Seoul, Korea
- 7Department of Laboratory Medicine, National Cancer Center, Goyang, Korea
- 8Department of Laboratory Medicine, Pusan National University School of Medicine, Yangsan, Korea
- KMID: 2525777
- DOI: http://doi.org/10.47429/lmo.2021.11.1.25
Abstract
- Background
The aim of this study was to investigate the status of next generation sequencing (NGS)-based genetic diagnosis in hematologic malignancies in Korea in 2017 and 2018.
Methods
A structured questionnaire was provided to specialists in charge of the genetic testing of hematologic malignancies via e-mail. The questionnaire consisted of 37 questions reflecting the situation of the institutions for each year and were based on an assessment of the status of the hematologic malignancy NGS test (19 questions) and the institution’s opinion on the NGS test (18 questions).
Results
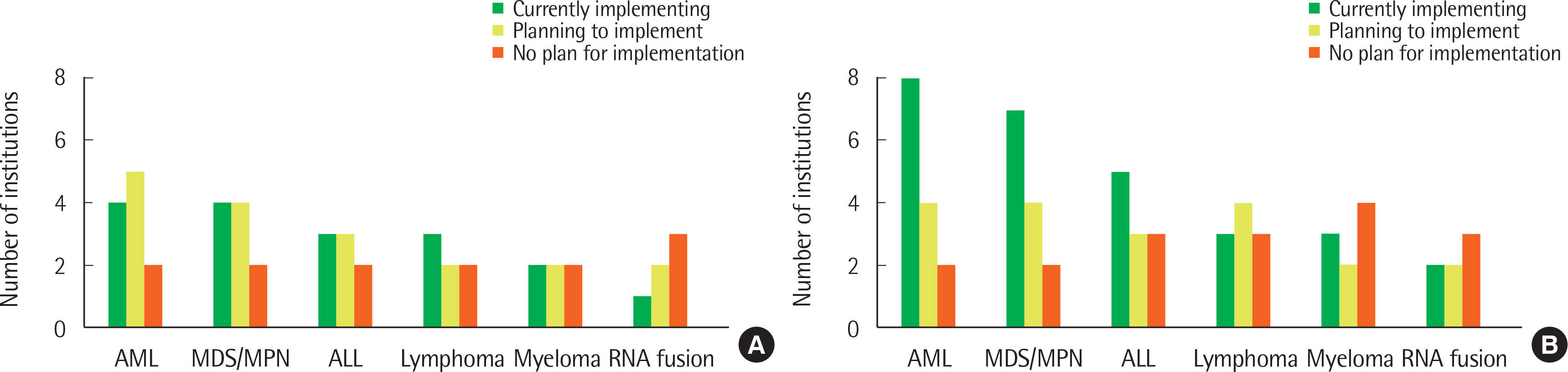
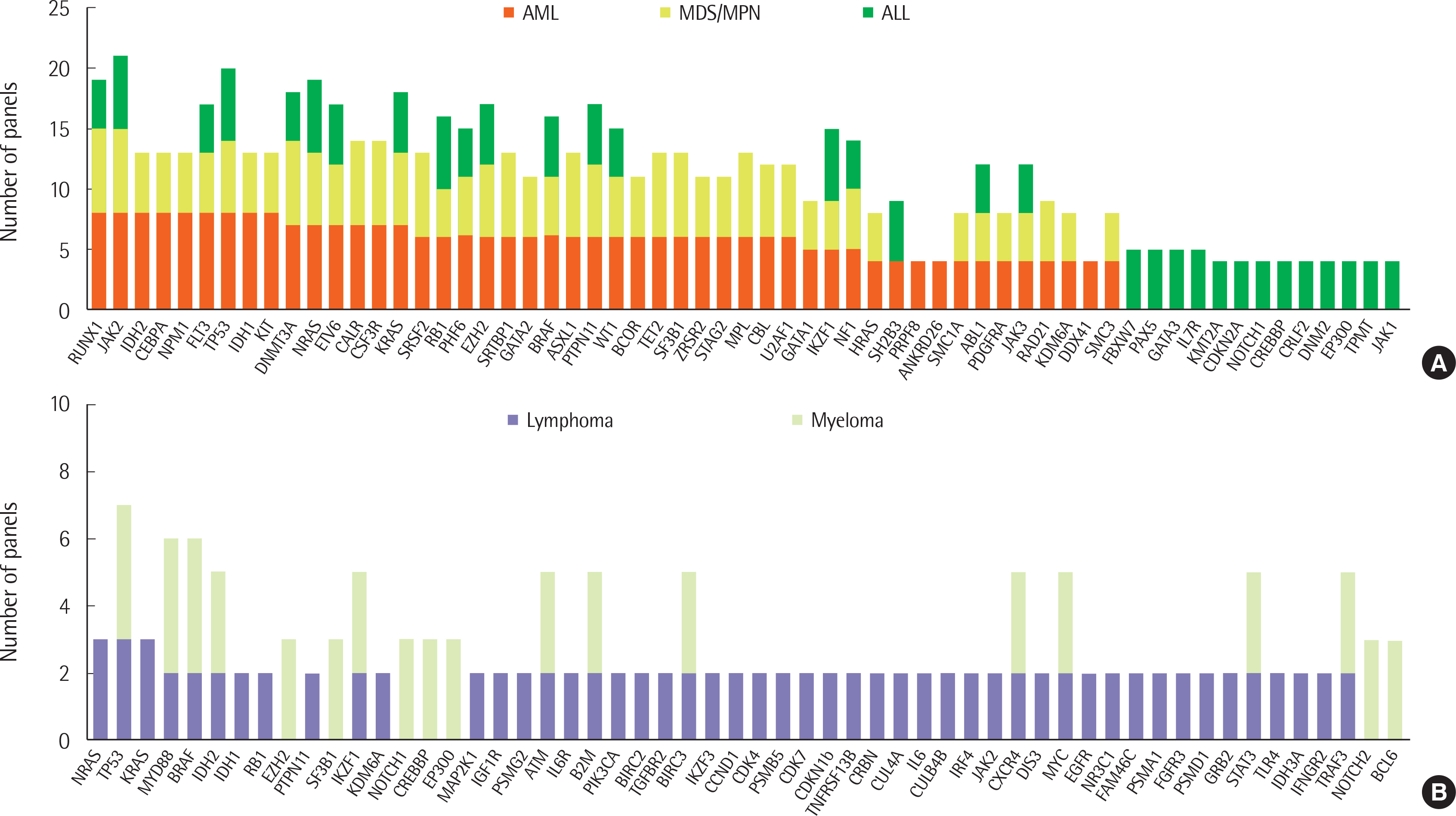
A total of 12 and 14 laboratories, in 2017 and 2018, respectively, replied to our survey and their answers were further analyzed. Most laboratories were performing NGS panel testing for acute leukemia and myeloid malignancies, and a small proportion of laboratories were testing NGS for lymphoid malignancies. The majority of participants agreed that NGS testing should be essential for the initial diagnostic workup.
Conclusions
Variation in NGS panel tests, including choice of gene and platform by different laboratories, were observed. Standardized panels and interpretation, centered around the Korean Society for Genetic Diagnostics, is needed to reduce inter-laboratory variation in NGS test results.
Keyword
Figure
Reference
-
1. Merker JD, Valouev A, Gotlib J. 2012; Next-generation sequencing in hematologic malignancies: what will be the dividends? Ther Adv Hematol. 3:333–9. DOI: 10.1177/2040620712458948. PMID: 23606936. PMCID: PMC3627325.2. Endrullat C, Glökler J, Franke P, Frohme M. 2016; Standardization and quality management in next-generation sequencing. Appl Transl Genom. 10:2–9. DOI: 10.1016/j.atg.2016.06.001. PMID: 27668169. PMCID: PMC5025460.3. Kim J, Park WY, Kim NKD, Jang SJ, Chun SM, Sung CO, et al. 2017; Good laboratory standards for clinical next-generation sequencing cancer panel tests. J Pathol Transl Med. 51:191–204. DOI: 10.4132/jptm.2017.03.14. PMID: 28535585. PMCID: PMC5445206.4. Mrózek K, Heerema NA, Bloomfield CD. 2004; Cytogenetics in acute leukemia. Blood Rev. 18:115–36. DOI: 10.1016/S0268-960X(03)00040-7.5. Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, et al. 2013; Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368:2059–74. DOI: 10.1056/NEJMoa1301689. PMID: 23634996. PMCID: PMC3767041.6. Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. 2013; Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 122:3616–27. DOI: 10.1182/blood-2013-08-518886. PMID: 24030381. PMCID: PMC3837510.7. Bacher U, Shumilov E, Flach J, Porret N, Joncourt R, Wiedemann G, et al. 2018; Challenges in the introduction of next-generation sequencing (NGS) for diagnostics of myeloid malignancies into clinical routine use. Blood Cancer J. 8:113. DOI: 10.1038/s41408-018-0148-6. PMID: 30420667. PMCID: PMC6232163.8. Aguilera-Diaz A, Vazquez I, Ariceta B, Mañú A, Blasco-Iturri Z, Palomino-Echeverría S, et al. 2020; Assessment of the clinical utility of four NGS panels in myeloid malignancies. Suggestions for NGS panel choice or design. PLoS One. 15:e0227986. DOI: 10.1371/journal.pone.0227986. PMID: 31978184. PMCID: PMC6980571.9. Thomas M, Sukhai MA, Zhang T, Dolatshahi R, Harbi D, Garg S, et al. 2017; Integration of technical, bioinformatic, and variant assessment appro-aches in the validation of a targeted next-generation sequencing panel for myeloid malignancies. Arch Pathol Lab Med. 141:759–75. DOI: 10.5858/arpa.2016-0547-RA. PMID: 28557600.10. Morgan GJ, Walker BA, Davies FE. 2012; The genetic architecture of multiple myeloma. Nat Rev Cancer. 12:335–48. DOI: 10.1038/nrc3257. PMID: 22495321.11. Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. 2015; Revised International Staging System for Multiple Myeloma: A report from International Myeloma Working Group. J Clin Oncol. 33:2863–9. DOI: 10.1200/JCO.2015.61.2267. PMID: 26240224. PMCID: PMC4846284.12. Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, et al. 2014; Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 28:384–90. DOI: 10.1038/leu.2013.199. PMID: 23817176. PMCID: PMC3916874.13. Kim B, Lee H, Shin S, Lee ST, Choi JR. 2019; Clinical evaluation of massively parallel RNA sequencing for detecting recurrent gene fusions in hematologic malignancies. J Mol Diagn. 21:163–70. DOI: 10.1016/j.jmoldx.2018.09.002. PMID: 30347268.14. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. 2016; Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 374:2209–21. DOI: 10.1056/NEJMoa1516192. PMID: 27276561. PMCID: PMC4979995.15. Feurstein S, Drazer MW, Godley LA. 2016; Genetic predisposition to leukemia and other hematologic malignancies. Semin Oncol. 43:598–608. DOI: 10.1053/j.seminoncol.2016.10.003. PMID: 27899193.16. Gao P, Zhang R, Li Z, Ding J, Xie J, Li J. 2019; Challenges of providing concordant interpretation of somatic variants in non-small cell lung cancer: A multicenter study. J Cancer. 10:1814–24. DOI: 10.7150/jca.29535. PMID: 31205538. PMCID: PMC6547979.17. National Institute of Food and Drug Safety Evaluation. 2018. 차세대염기서열분석(Next Generation Sequencing) 임상검사실 인증 검사분야별 가이드라인체세포(Somatic). National Institute of Food and Drug Safety Evaluation.18. Kim H, Yun JW, Lee ST, Kim HJ, Kim SH, Kim JW, et al. 2019; Korean Society for Genetic Diagnostics Guidelines for Validation of Next-generation Sequencing-based Somatic Variant Detection in Hematologic Malignancies. Ann Lab Med. 39:515–23. DOI: 10.3343/alm.2019.39.6.515. PMID: 31240878. PMCID: PMC6660343.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Korean Society for Genetic Diagnostics Guidelines for Validation of Next-Generation Sequencing-Based Somatic Variant Detection in Hematologic Malignancies
- Principles of Genetic Counseling in the Era of Next-Generation Sequencing
- Recent Advances in the Clinical Application of Next-Generation Sequencing
- Ultra-rare Disease and Genomics-Driven Precision Medicine
- Comprehensive Molecular Characterization of Urological Malignancies: Literature Review of Landmark Studies