Ann Lab Med.
2021 Jan;41(1):86-94. 10.3343/alm.2021.41.1.86.
Antimicrobial Resistance Caused by KPC-2 Encoded by Promiscuous Plasmids of the Klebsiella pneumoniae ST307 Strain
- Affiliations
-
- 1Department of Microbiology, Pukyoung National University, Busan, Korea
- 2Department of Laboratory Medicine, BHS Hanseo Hospital, Busan, Korea
- KMID: 2512737
- DOI: http://doi.org/10.3343/alm.2021.41.1.86
Abstract
- Background
A lineage of Klebsiella pneumoniae that produces carbapenemase-2 (KPC-2), sequence type (ST) 307, emerged in 2017. We analyzed the complete sequences of plasmids from KPC-2-producing K. pneumoniae (KPC-Kp) ST307, investigated the antimicrobial resistance conferred by this strain, and confirmed the horizontal interspecies transmission of KPC- carbapenemase-producing Enterobacteriaceae (CPE) characteristics among Enterobacteriaceae.
Methods
We performed antimicrobial susceptibility testing, PCR analysis, multilocus sequence typing, curing tests, and whole-genome sequencing to characterize plasmid-derived KPC-2-producing Enterobacteriaceae clinical isolates.
Results
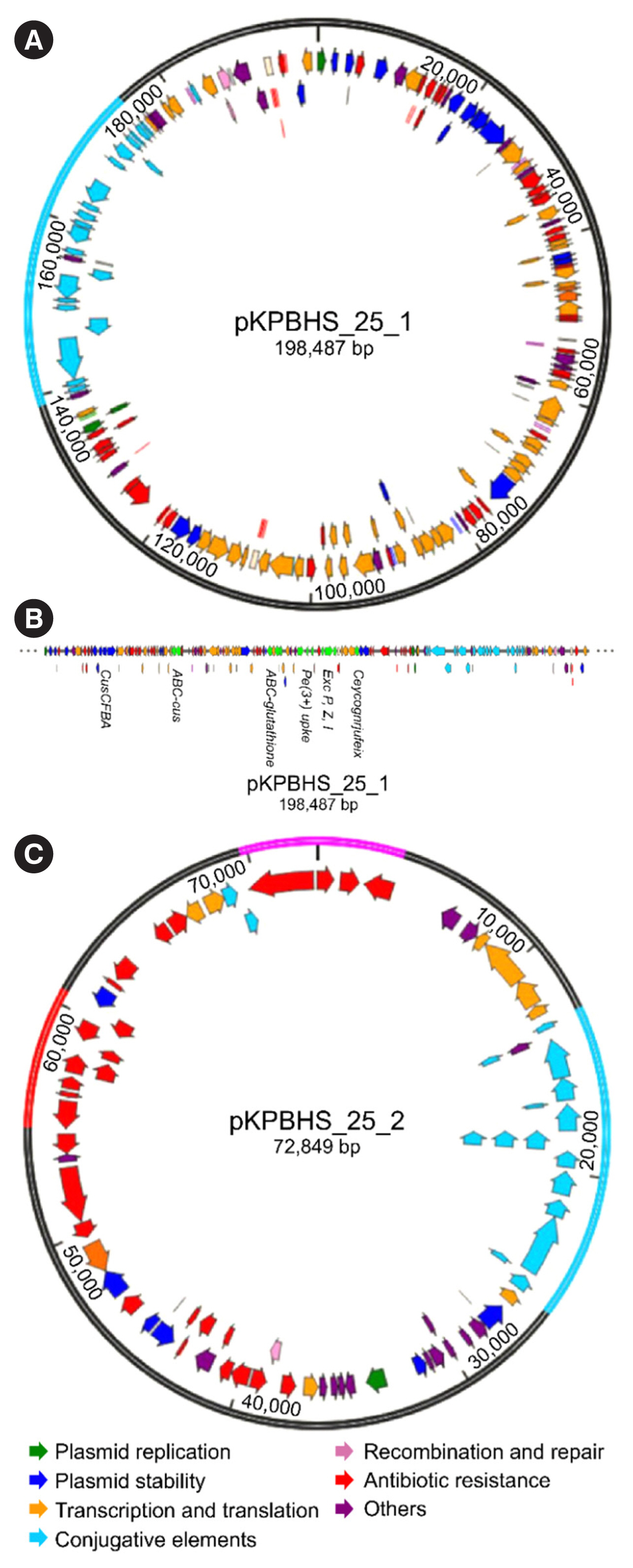
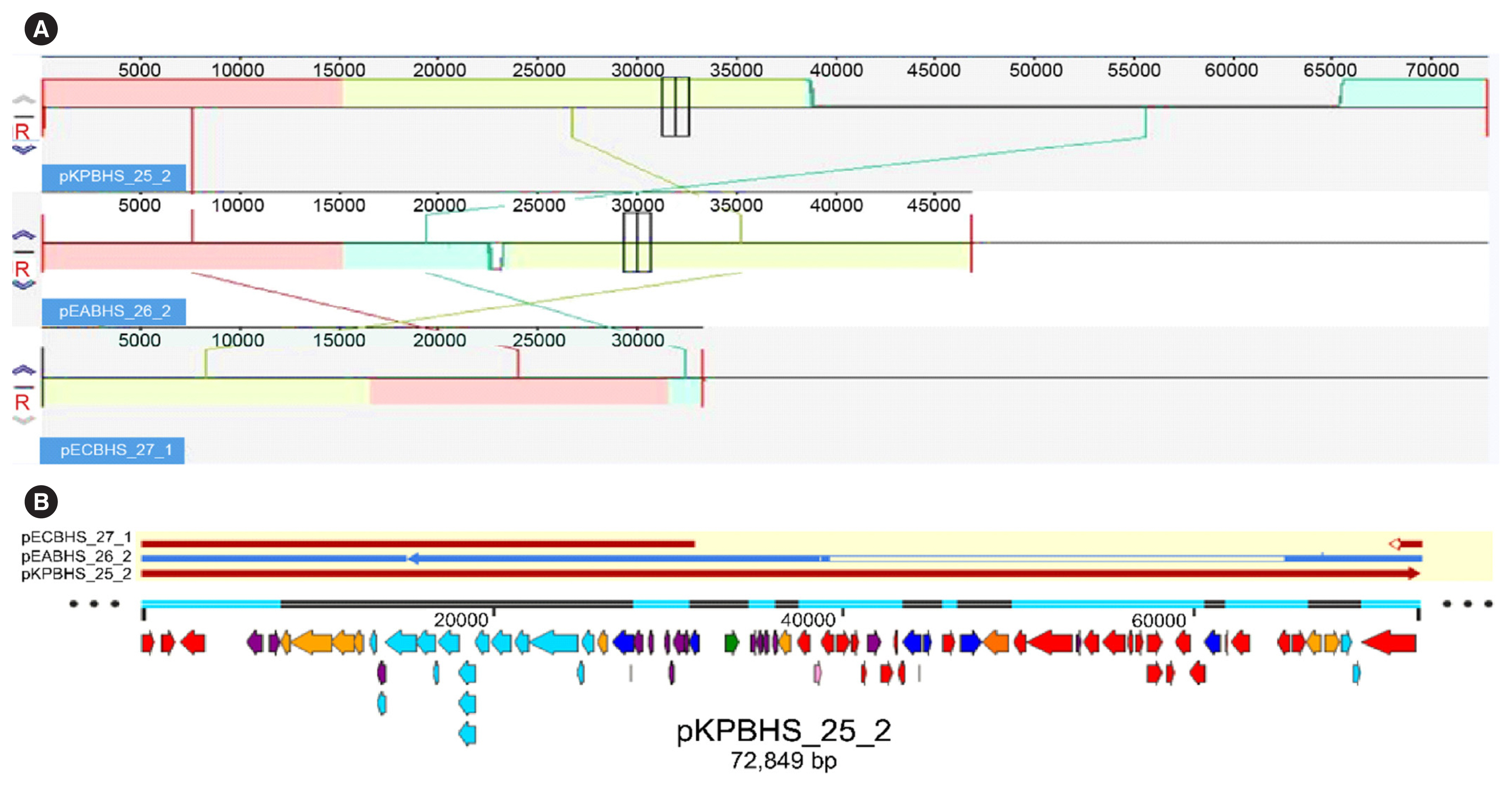
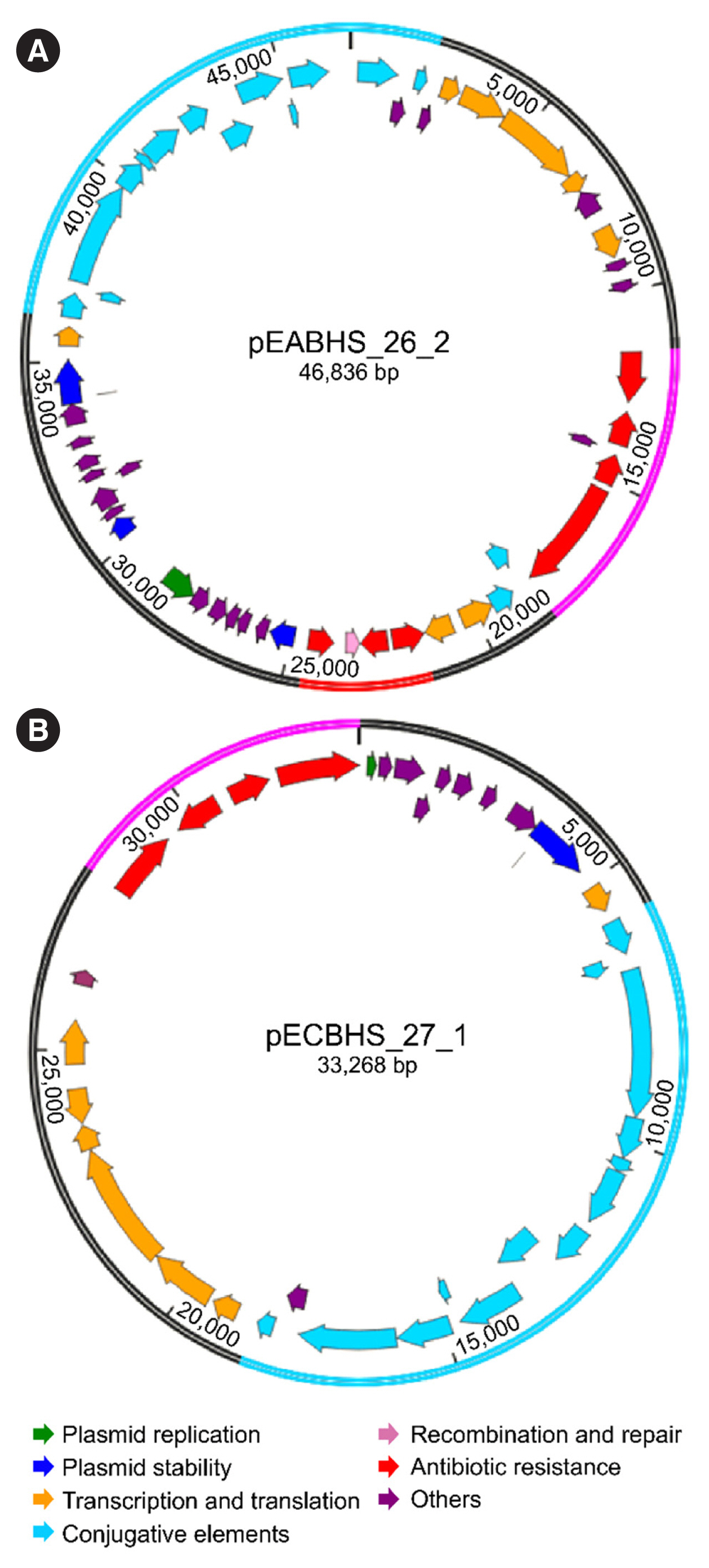
Sequence analysis of KPC-Kp strain ST307 revealed novel plasmid-located virulence factors, including a gene cluster for glycogen synthesis. Three Enterobacteriaceae strains were identified in one patient: K. pneumoniae (CPKp1825), Klebsiella aerogenes (CPEa1826), and Escherichia coli (CPEc1827). The bla KPC-2 gene from K. pneumoniae ST307 was horizontally transmitted between these strains. The plasmids could be transferred through conjugation, because all three strains of bacteria contained the type IV secretion system, pilus genes, and tra genes for conjugal transfer. The bla KPC-2 gene was located on a truncated Tn4401 transposon. Plasmids containing the bla KPC-2 gene could not be artificially removed; thus, the three strains could not be cured.
Conclusions
The ease of horizontal transfer of KPC-Kp ST307 carbapenem resistance has serious public health and epidemiological implications. This study provides a better understanding of the genetic characteristics that can contribute to the growth and spread of KPC-Kp ST307, and their association with antimicrobial resistance genes.
Keyword
Figure
Reference
-
1. Yoon EJ, Oh Y, Jeong SH. Development of tigecycline resistance in carbapenemase-producing Klebsiella pneumoniae sequence type 147 via AcrAB overproduction mediated by replacement of the ramA promoter. Ann Lab Med. 2020; 40:15–20.2. O’Neill J. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. London: Review of antimicrobial resistance;https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (Updated on Dec 2016).3. Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015; 59:5873–84.4. Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, et al. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci U S A. 2014; 111:4988–93.5. Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016; 24:944–56.6. Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014; 20:350–4.7. Castanheira M, Farrell SE, Wanger A, Rolston KV, Jones RN, Mendes RE. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb Drug Resist. 2013; 19:295–7.8. Ocampo AM, Chen L, Cienfuegos AV, Roncancio G, Chavda KD, Kreiswirth BN, et al. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother. 2015; 60:332–42.9. He S, Chandler M, Varani AM, Hickman AB, Dekker JP, Dyda F. Mechanisms of evolution in high-consequence drug resistance plasmids. MBio. 2016; 7:e01987–16.
Article10. Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016; 7:895.
Article11. Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014; 69:628–31.12. Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012; 68:43–50.13. Cicek AC, Duzgun AO, Saral A, Sandalli C. Determination of a novel integron-located variant (blaOXA-320) of class D β-lactamase in Proteus mirabilis. J Basic Microbiol. 2014; 54:1030–5.14. CLSI. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 11th ed. CLSI M07. Wayne, PA: Clinical and Laboratory Standards Institute;2018.15. Jeong S, Kim JO, Jeong SH, Bae IK, Song W. Evaluation of peptide nucleic acid-mediated multiplex real-time PCR kits for rapid detection of carbapenemase genes in gram-negative clinical isolates. J Microbiol Methods. 2015; 113:4–9.
Article16. Landman D, Bratu S, Quale J. Contribution of ompK36 to carbapenem susceptibility in KPC-producing Klebsiella pneumoniae. J Med Microbiol. 2009; 58:1303–8.17. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005; 43:4178–82.18. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006; 60:1136–51.19. Jeong SH, Lee KM, Lee J, Bae IK, Kim JS, Kim HS, et al. Clonal and horizontal spread of the blaOXA-232 gene among Enterobacteriaceae in a Korean hospital. Diagn Microbiol Infect Dis. 2015; 82:70–2.20. Leavitt A, Chmelnitsky I, Ofek I, Carmeli Y, Navon-Venezia S. Plasmid pKpQIL encoding KPC-3 and TEM-1 confers carbapenem resistance in an extremely drug-resistant epidemic Klebsiella pneumoniae strain. J Antimicrob Chemother. 2010; 65:243–8.21. Yoon EJ, Kim JO, Kim D, Lee H, Yang JW, Lee KJ, et al. Klebsiella pneumoniae carbapenemase producers in South Korea between 2013 and 2015. Front Microbiol. 2018; 9:56.
Article22. Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, et al. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One. 2015; 10:e0132936.23. Yoon EJ, Yang JW, Kim JO, Lee H, Lee KJ, Jeong SH. Carbapenemase-producing Enterobacteriaceae in South Korea: A report from the National Laboratory Surveillance System. Future Microbiol. 2018; 13:771–83.24. Long SW, Olsen RJ, Eagar TN, Beres SB, Zhao P, Davis JJ, et al. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: Unexpected abundance of clonal group 307. mBio. 2017; 8:e00489–17.
Article25. Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017; 3:e000110.
Article26. Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother. 2019; 74:577–81.27. Roth AL, Kurpiel PM, Lister PD, Hanson ND. blaKPC RNA expression correlates with two transcriptional start sites but not always with gene copy number in four genera of gram-negative pathogens. Antimicrob Agents Chemother. 2011; 55:3936–8.28. Tsai YK, Fung CP, Lin JC, Chen JH, Chang FY, Chen TL, et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2011; 55:1485–93.29. Kim SY, Park YJ, Yu JK, Kim YS. Aminoglycoside susceptibility profiles of Enterobacter cloacae isolates harboring the aac (6′)-Ib gene. Korean J Lab Med. 2011; 31:279–81.30. Leclercq R, Cantón R, Brown DF, Giske CG, Heisig P, MacGowan AP, et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin Microbiol Infect. 2013; 19:141–60.
Article31. Almaghrabi R, Clancy CJ, Doi Y, Hao B, Chen L, Shields RK, et al. Carbapenem-resistant Klebsiella pneumoniae strains exhibit diversity in aminoglycoside-modifying enzymes, which exert differing effects on plazomicin and other agents. Antimicrob Agents Chemother. 2014; 58:4443–51.32. Jakobsen L, Sandvang D, Jensen VF, Seyfarth AM, Frimodt-M⊘ller N, Hammerum AM. Gentamicin susceptibility in Escherichia coli related to the genetic background: problems with breakpoints. Clin Microbiol Infect. 2007; 13:830–2.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MCR1 and KPC2 Co-producing Klebsiella pneumoniae Bacteremia: First Case in Korea
- Prevalence and antimicrobial resistance of Klebsiella species isolated from clinically ill companion animals
- Isolation of a Klebsiella pneumoniae Isolate of Sequence Type 258 Producing KPC-2 Carbapenemase in Korea
- Epidemiological Study of an Outbreak of KPC-2-producing Klebsiella pneumoniae in a Tertiary Hospital in Korea
- Association Between Toxin-antitoxin Systems on Plasmids and Persister Formation in CTX-15-producing Klebsiella pneumoniae ST11 Isolates