Ann Lab Med.
2021 May;41(3):350-353. 10.3343/alm.2021.41.3.350.
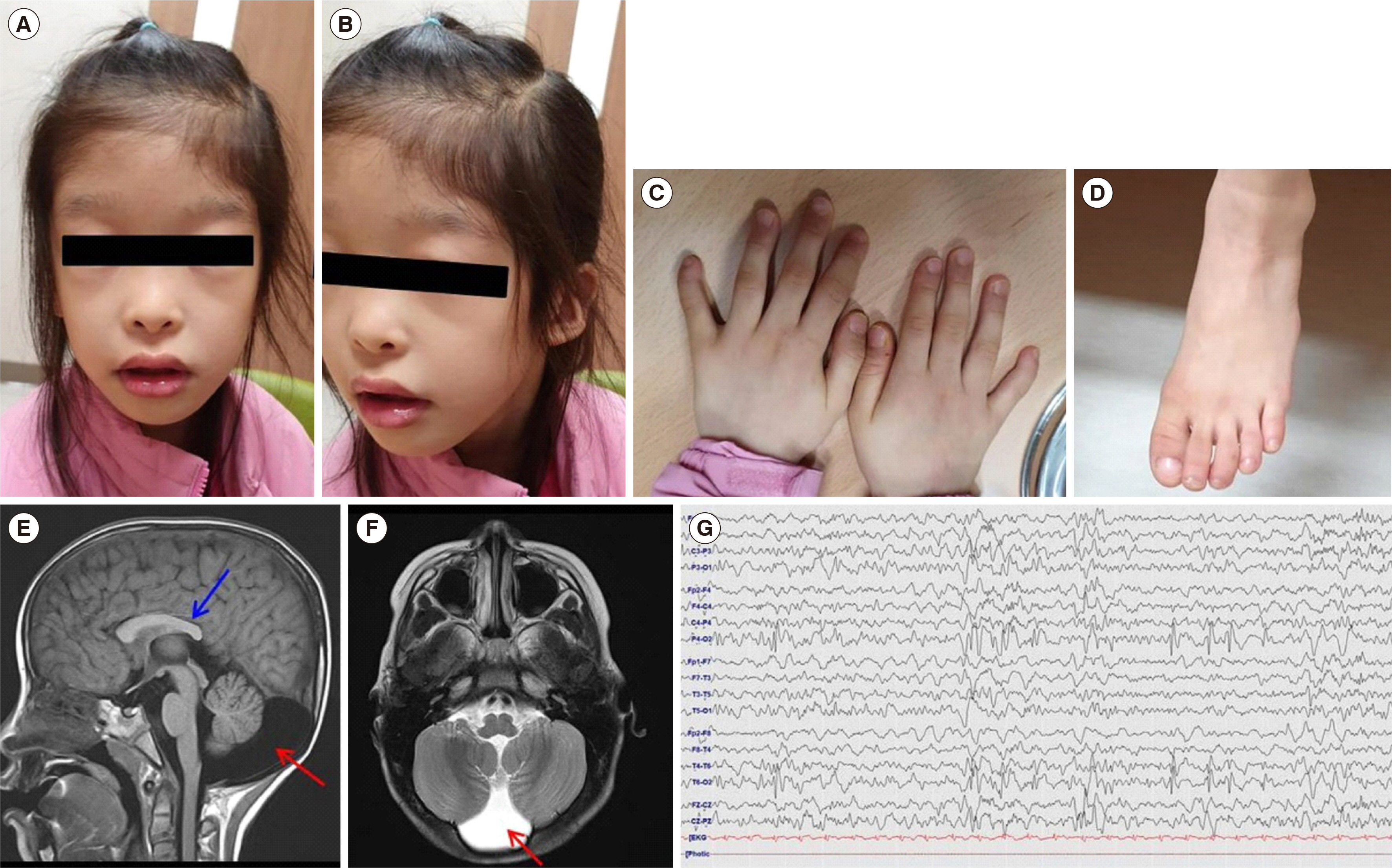
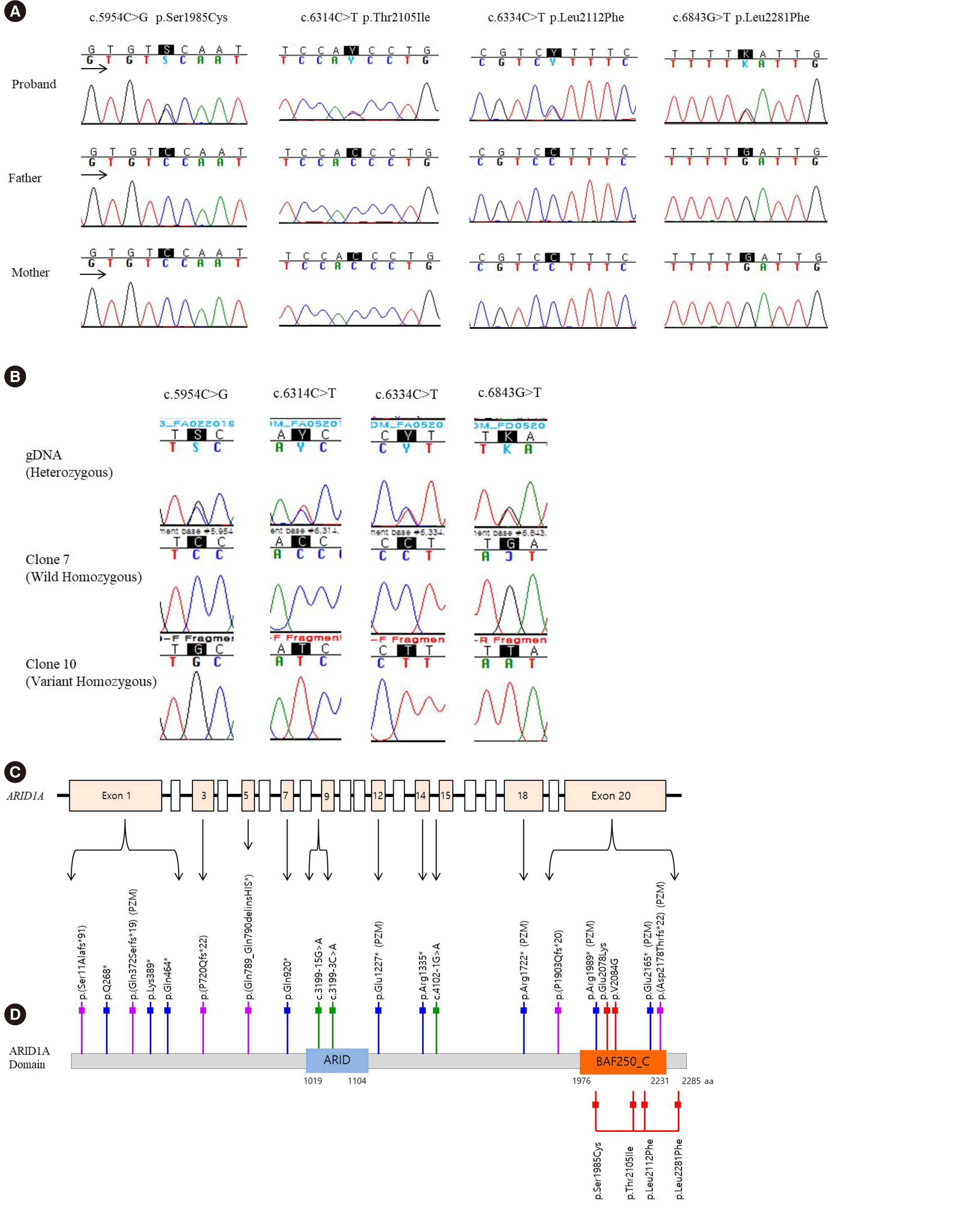
A Novel De Novo Heterozygous ARID1A Missense Variant Cluster in cis c.[5954C>G;6314C>T;6334C>T; 6843G>C] causes a Coffin–Siris Syndrome
- Affiliations
-
- 1Department of Pediatrics, Nowon Eulji Medical Center, Eulji University, Seoul, Korea
- 2GC Genome, Yongin, Korea
- KMID: 2512698
- DOI: http://doi.org/10.3343/alm.2021.41.3.350
Figure
Reference
-
1. Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, Hibi-Ko Y, et al. 2012; Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 44:376–8. DOI: 10.1038/ng.2219. PMID: 22426308.
Article2. Sekiguchi F, Tsurusaki Y, Okamoto N, Teik KW, Mizuno S, Suzumura H, et al. 2019; Genetic abnormalities in a large cohort of Coffin-Siris syndrome patients. J Hum Genet. 64:1173–86. DOI: 10.1038/s10038-019-0667-4. PMID: 31530938.3. Santen GW, Aten E, Vulto-van Silfhout AT, Pottinger C, van Bon BW, van Minderhout IJ, et al. 2013; Coffin-Siris syndrome and the BAF complex: genotype-phenotype study in 63 patients. Hum Mutat. 34:1519–28. DOI: 10.1002/humu.22394. PMID: 23929686.
Article4. Martínez F, Caro-Llopis A, Roselló M, Oltra S, Mayo S, Monfort S, et al. 2017; High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J Med Genet. 54:87–92. DOI: 10.1136/jmedgenet-2016-103964. PMID: 27620904.
Article5. Dillon OJ, Lunke S, Stark Z, Yeung A, Thorne N, et al. Melbourne Genomics Health Alliance. 2018; Exome sequencing has higher diagnostic yield compared to simulated disease-specific panels in children with suspected monogenic disorders. Eur J Hum Genet. 26:644–51. DOI: 10.1038/s41431-018-0099-1. PMID: 29453417. PMCID: PMC5945679.
Article6. Wieczorek D, Bögershausen N, Beleggia F, Steiner-Haldenstätt S, Pohl E, Li Y, et al. 2013; A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 22:5121–35. DOI: 10.1093/hmg/ddt366. PMID: 23906836.
Article7. Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Tippin Davis B, et al. 2015; Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 17:578–86. DOI: 10.1038/gim.2014.154. PMID: 25356970.
Article8. Kwak KJ, Oh SW, Kim CT. 2011. Korean-Wechsler Intelligence Scale for Children. 4th ed. Hakjisa;Seoul: p. 11–3.9. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. 2015; Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–24. DOI: 10.1038/gim.2015.30. PMID: 25741868. PMCID: PMC4544753.
Article10. Karbassi I, Maston GA, Love A, DiVincenzo C, Braastad CD, Elzinga CD, et al. 2016; A standardized DNA variant scoring system for pathogenicity assessments in mendelian disorders. Hum Mutat. 37:127–34. DOI: 10.1002/humu.22918. PMID: 26467025. PMCID: PMC4737317.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Coffin-Siris Syndrome: Genotype-Phenotype Clustering and Novel Variants
- Identification of a likely pathogenic variant of YY1 in a patient with developmental delay
- De novo HCN1 Mutation Identified by Next-Generation Sequencing in a Patient with Early Infantile Epileptic Encephalopathy: Case Report
- Identification of a De Novo Heterozygous Missense FLNB Mutation in Lethal Atelosteogenesis Type I by Exome Sequencing
- Identification of a Novel De Novo Variant in the PAX3 Gene in Waardenburg Syndrome by Diagnostic Exome Sequencing: The First Molecular Diagnosis in Korea