Intest Res.
2021 Jan;19(1):12-20. 10.5217/ir.2019.00133.
Two intertwined compartments coexisting in sporadic conventional colon adenomas
- Affiliations
-
- 1Gastrointestinal Research Laboratory, Department of Pathology, Karolinska Institute and University Hospital, Stockholm, Sweden
- KMID: 2512097
- DOI: http://doi.org/10.5217/ir.2019.00133
Abstract
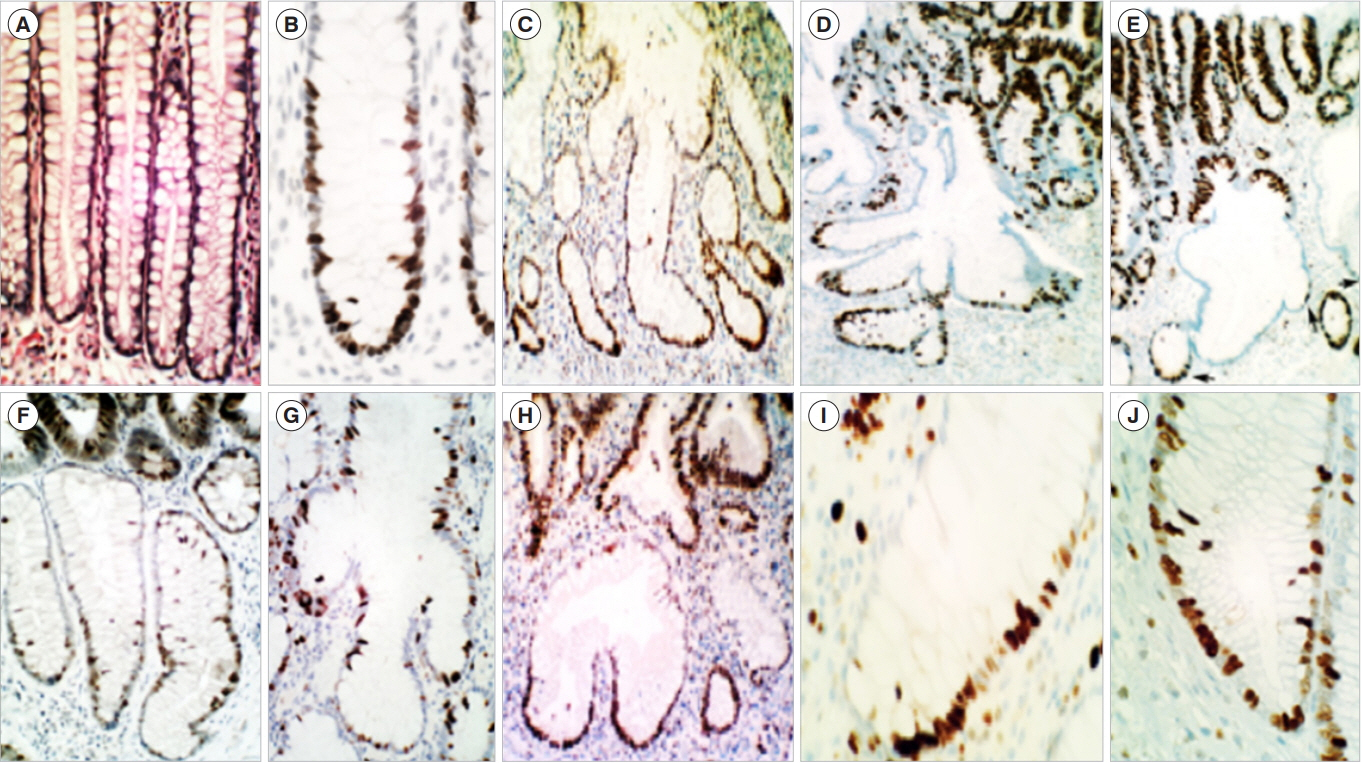
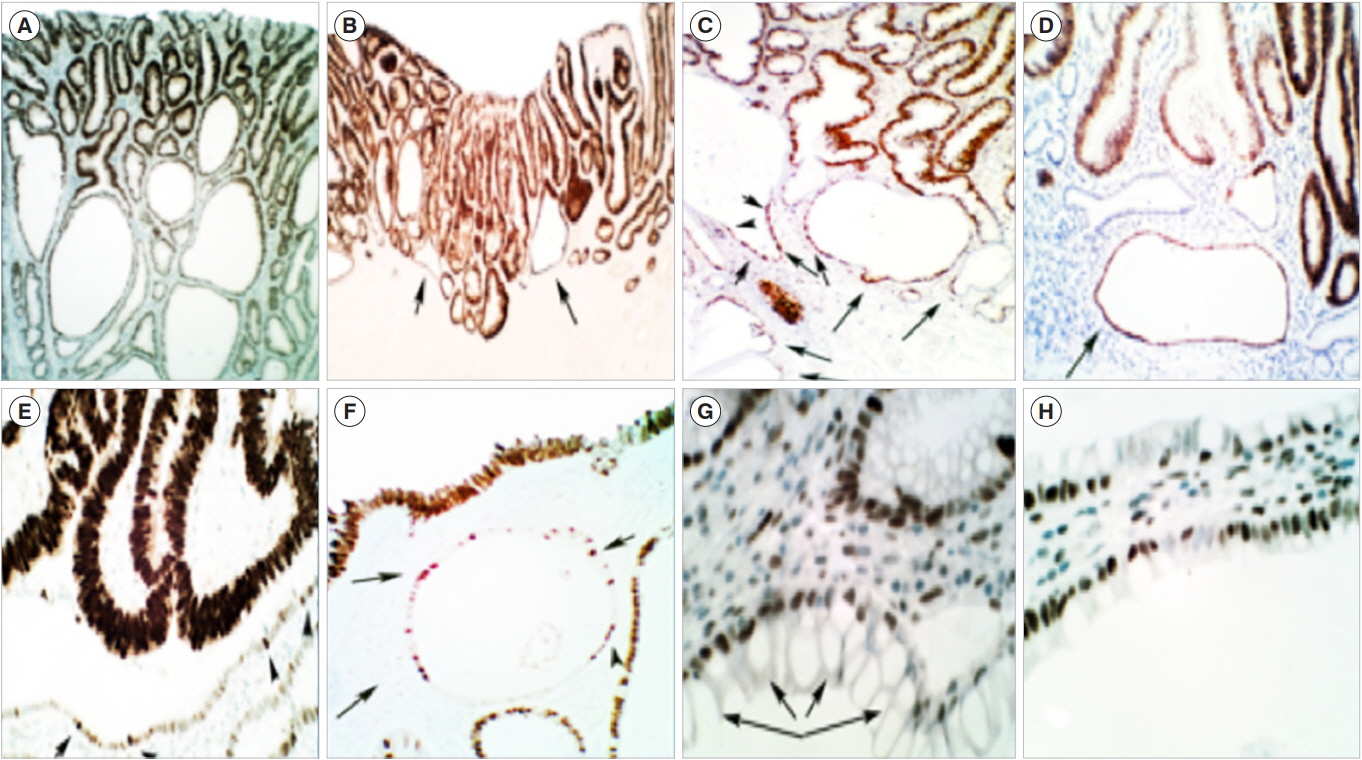
- Sporadic conventional colon adenomas are microscopically built of 2 intertwined compartments: one on top, harboring the dysplastic tissue that defines their histo-biomolecular attributes, and the other below, composed of non-dysplastic crypts with corrupted shapes (CCS). The CCS of 306 colon adenomas revealed asymmetric, haphazardly-distributed proliferating cell-domains (PC). In contrast, the PC-domains in normal controls were symmetric, being limited to the lower thirds of the crypts. In 28% out of 501 sporadic conventional adenomas, foci of p53-upregulated dysplastic tissue were found. The CCS in 30% of 108 sporadic adenomas showed p53-upregulated single cells, suggesting mounting somatic mutations. No p53-upregulated cells were found in the crypts of controls. In polypoid adenomas, the mucosa of the stalk without dysplastic tissue on top disclosed CCS with asymmetrical PC-domains and single p53-upregulated cells. The latter observations suggested that CCS had developed prior to and not after the growth of the dysplastic tissue on top. CCS were also found below colon adenomas in carcinogen-treated rats. It is concluded that the 2 intertwined histo-biological compartments of sporadic conventional colon adenomas are probably interdependent components. These findings may open new directions aimed to uncover the link between the normal colonic mucosa and the histogenesis of, conventional adenomas.
Figure
Cited by 3 articles
-
Gut Microbiome and Colorectal Cancer
Tae-Geun Gweon
Korean J Gastroenterol. 2023;82(2):56-62. doi: 10.4166/kjg.2023.089.Calcium, Vitamin D, and Colorectal Cancer
Young-Jo Wi, Soo-Young Na
Korean J Gastroenterol. 2023;82(2):47-55. doi: 10.4166/kjg.2023.091.Summary and comparison of recently updated post-polypectomy surveillance guidelines
Yoon Suk Jung
Intest Res. 2023;21(4):443-451. doi: 10.5217/ir.2023.00107.
Reference
-
1. Dahl J, Greenson JK. Colon. In : Mills SE, editor. Histology for pathologists. 3rd ed. Philadelphia: Lippincott Williams & Wilkins;2007. p. 627–648.2. Baker AM, Gabbutt C, Williams MJ, et al. Crypt fusion as a homeostatic mechanism in the human colon. Gut. 2019; 68:19861993.
Article3. Boman BM, Fields JZ. An APC:WNT counter-current-like mechanism regulates cell division along the human colonic crypt axis: a mechanism that explains how APC mutations induce proliferative abnormalities that drive colon cancer development. Front Oncol. 2013; 3:244.
Article4. Rubio CA. The histologic structure of the large bowel mucosa and the evolution of the three pathways of colonic carcinogenesis in humans and in experimental animals. Recent Stud Dig Syst Anat. 2018; 3:1–12.5. Levine DS, Haggitt RC. Normal histology of the colon. Am J Surg Pathol. 1989; 13:966–984.
Article6. Senior PV, Pritchett CJ, Sunter JP, Appleton DR, Watson AJ. Crypt regeneration in adult human colonic mucosa during prolonged organ culture. J Anat. 1982; 134(Pt 3):459–469.7. Rubio CA. Are corrupted non-dysplastic colonic crypts the first histological event in experimental colonic carcinogenesis? Anticancer Res. 2017; 37:2265–2268.
Article8. Helander HF, Fändriks L. Surface area of the digestive tract: revisited. Scand J Gastroenterol. 2014; 49:681–689.9. Reddy Parine N, Alanazi IO, Shaik JP, et al. TDG gene polymorphisms and their possible association with colorectal cancer: a case control study. J Oncol. 2019; 2019:7091815.
Article10. Davenport JR, Su T, Zhao Z, et al. Modifiable lifestyle factors associated with risk of sessile serrated polyps, conventional adenomas and hyperplastic polyps. Gut. 2018; 67:456–465.
Article11. Deb M, Sengupta D, Kar S, et al. Epigenetic drift towards histone modifications regulates CAV1 gene expression in colon cancer. Gene. 2016; 581:75–84.
Article12. O’Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016; 13:691–706.
Article13. Seidel DV, Azcárate-Peril MA, Chapkin RS, Turner ND. Shaping functional gut microbiota using dietary bioactives to reduce colon cancer risk. Semin Cancer Biol. 2017; 46:191–204.
Article14. Rubio CA, Langner C, Schmidt PT. Partial to complete abrogation of the subepithelial macrophage barrier against the gut microbiota in patients with ulcerative colitis and Crohn’s colitis. Histopathology. 2018; 72:580–587.
Article15. Bailie L, Loughrey MB, Coleman HG. Lifestyle risk factors for serrated colorectal polyps: a systematic review and meta-analysis. Gastroenterology. 2017; 152:92–104.
Article16. Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett. 2018; 16:9–18.17. Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014; 27:9–14.18. Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel). 2013; 5:676–713.
Article19. Bargen JA. Rectal and sigmoidal adenomas: potential carcinoma. Gastroenterology. 1951; 18:138–140.20. Hamilton SR, Vogelstein B, Kudo S, et al. Carcinoma of the colon and rectum. In : Hamilton SR, Aaltonen LA, editors. World Health Organization Classification of Tumours. Pathology and genetics of tumours of the digestive system. Lyon: IARC Press;2000. p. 104–119.21. Schlemper RJ, Riddell RH, Kato Y, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000; 47:251255.
Article22. Komor MA, Bosch LJ, Bounova G, et al. Consensus molecular subtype classification of colorectal adenomas. J Pathol. 2018; 246:266–276.
Article23. Sakai E, Fukuyo M, Ohata K, et al. Genetic and epigenetic aberrations occurring in colorectal tumors associated with serrated pathway. Int J Cancer. 2016; 138:1634–1644.
Article24. Neuville A, Nicolet C, Meyer N, et al. Histologic characteristics of non-microsatellite-instable colon adenomas correlate with distinct molecular patterns. Hum Pathol. 2011; 42:244–253.
Article25. Jackman RJ, Mayo CW. The adenoma-carcinoma sequence in cancer of the colon. Surg Gynecol Obstet. 1951; 93:327–330.26. Longacre TA, Fenoglio-Preiser CM. Mixed hyperplastic adenomatous polyps/serrated adenomas: a distinct form of colorectal neoplasia. Am J Surg Pathol. 1990; 14:524–537.27. Torlakovic EE, Gomez JD, Driman DK, et al. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA). Am J Surg Pathol. 2008; 32:21–29.
Article28. O’Brien MJ, Gibbons D. The adenoma-carcinoma sequence in colorectal neoplasia. Surg Oncol Clin N Am. 1996; 5:513–530.
Article29. O’Brien MJ, Zhao Q, Yang S. Colorectal serrated pathway cancers and precursors. Histopathology. 2015; 66:49–65.
Article30. Rubio CA. Colorectal adenomas: time for reappraisal. Pathol Res Pract. 2002; 198:615–620.
Article31. Rubio CA, Kis L, Nastic D, Schmidt PT. Microtubular colorectal adenoma an aggressive histologic phenotype with propensity to evolve into invasive carcinoma. ARC J Cancer Sci. 2017; 3:31–36.32. Muto T, Kamiya J, Sawada T, et al. Small “flat adenoma” of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum. 1985; 28:847–851.
Article33. Kasumi A, Kratzer GL, Takeda M. Observations of aggressive, small, flat, and depressed colon cancer: report of three cases. Surg Endosc. 1995; 9:690–694.
Article34. Kudo S, Kashida H, Tamura S, Nakajima T. The problem of “flat” colonic adenoma. Gastrointest Endosc Clin N Am. 1997; 7:8798.
Article35. Kudo Se, Lambert R, Allen JI, et al. Nonpolypoid neoplastic lesions of the colorectal mucosa. Gastrointest Endosc. 2008; 68(4 Suppl):S3–S47.
Article36. Bird RP. Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: preliminary findings. Cancer Lett. 1987; 37:147–151.
Article37. Nascimbeni R, Villanacci V, Mariani PP, et al. Aberrant crypt foci in the human colon: frequency and histologic patterns in patients with colorectal cancer or diverticular disease. Am J Surg Pathol. 1999; 23:1256–1263.38. Drew DA, Mo A, Grady JJ, et al. Proximal aberrant crypt foci associate with synchronous neoplasia and are primed for neoplastic progression. Mol Cancer Res. 2018; 16:486–495.
Article39. Fenoglio-Preiser CM, Noffsinger A. Aberrant crypt foci: a review. Toxicol Pathol. 1999; 27:632–642.40. Dolara P, Caderni G, Luceri C. Surrogate endpoint biomarkers for human colon carcinogenesis. Toxicol Lett. 2000; 112–-113:415420.
Article41. Ochiai M, Hippo Y, Izumiya M, Watanabe M, Nakagama H. Newly defined aberrant crypt foci as a marker for dysplasia in the rat colon. Cancer Sci. 2014; 105:943–950.
Article42. Quintanilla I, López-Cerón M, Jimeno M, et al. Rectal aberrant crypt foci in humans are not surrogate markers for colorectal cancer risk. Clin Transl Gastroenterol. 2019; 10:e00047.
Article43. Rubio CA, Schmidt PT. Are non-dysplastic crypts with corrupted shapes the initial recordable histological event in the development of sporadic conventional adenomas? Anticancer Res. 2018; 38:5315–5320.
Article44. Rubio CA, Schmidt PT. Crypts with corrupted shapes in nonpolypoid adenomas. Anticancer Res. 2019; 39:833–838.
Article45. Rubio CA. Preliminary report: multiple clusters of proliferating cells in non-dysplastic corrupted colonic crypts underneath conventional adenomas. In Vivo. 2018; 32:1473–1475.
Article46. Rubio CA, Schmidt PT. The normal epithelium of crypts accruing below nonpolypoid adenomas thrives with relocated proliferating cell-domains and p53-up-regulated cells. Anticancer Res. 2019; 39:4965–4970.
Article47. Maskens AP, Deschner EE. Tritiated thymidine incorporation into epithelial cells of normal-appearing colorectal mucosa of cancer patients. J Natl Cancer Inst. 1977; 58:1221–1224.
Article48. Arai T, Kino I. Morphometrical and cell kinetic studies of normal human colorectal mucosa: comparison between the proximal and the distal large intestine. Acta Pathol Jpn. 1989; 39:725–730.
Article49. Gonzalez RS, Messing S, Tu X, McMahon LA, Whitney-Miller CL. Immunohistochemistry as a surrogate for molecular subtyping of gastric adenocarcinoma. Hum Pathol. 2016; 56:16–21.
Article50. Rubio CA, Schmidt PT. Disparate cell proliferation and p53 overexpression in colonic crypts with normal epithelial lining found below the neoplastic canopy of conventional adenomas. J Pathol Clin Res. 2019; 5:154–163.
Article51. Rubio CA, Rodesjö M, Duvander A, Mathies M, Garberg L, Shetye J. p53 up-regulation during colorectal carcinogenesis. Anticancer Res. 2014; 34:6973–6979.52. Xu J, Galley JD, Bailey MT, Thomas-Ahner JM, Clinton SK, Olivo-Marston SE. The impact of dietary energy intake early in life on the colonic microbiota of adult mice. Sci Rep. 2016; 6:19083.
Article53. Zhou L, Zahid M, Anwar MM, et al. Suggestive evidence for the induction of colonic aberrant crypts in mice fed sodium nitrite. Nutr Cancer. 2016; 68:105–112.
Article54. Kristiansen E. The role of aberrant crypt foci induced by the two heterocyclic amines 2-amino-3-methyl-imidazo[4,5-f] quinoline (IQ) and 2-amino-1-methyl-6-phenyl-imidazo[4,5b]pyridine (PhIP) in the development of colon cancer in mice. Cancer Lett. 1996; 110:187–192.
Article55. Ghirardi M, Nascimbeni R, Villanacci V, Fontana MG, Di Betta E, Salerni B. Azoxymethane-induced aberrant crypt foci and colorectal tumors in F344 rats: sequential analysis of growth. Eur Surg Res. 1999; 31:272–280.
Article56. Richards TC. Early changes in the dynamics of crypt cell populations in mouse colon following administration of 1,2-dimethylhydrazine. Cancer Res. 1977; 37:1680–1685.57. Sunter JP, Wright NA, Appleton DR. Cell population kinetics in the epithelium of the colon of the male rat. Virchows Arch B Cell Pathol. 1978; 26:275–287.
Article58. Wasan HS, Park HS, Liu KC, et al. APC in the regulation of intestinal crypt fission. J Pathol. 1998; 185:246–255.
Article59. Rubio CA. Corrupted colonic crypt fission in carcinogen-treated rats. PLoS One. 2017; 12:e0172824.
Article60. Rubio CA. Putative stem cells in mucosas of the esophago-gastrointestinal tract. In : Singh SR, editor. Stem cell, regenerative medicine and cancer. New York: Nova Science Publishers Inc;2010. p. 279–308.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Hyperplastic Polyposis Associated with Two Synchronous Colon Cancers
- Endoscopic Resection of an Adenocarcinoma Arising from a Sporadic Tubulovillous Adenoma of the Duodenum
- Non-conventional dysplastic subtypes in inflammatory bowel disease: a review of their diagnostic characteristics and potential clinical implications
- Sigmoidoscopy, is it Enough as a Screening Tool? -Undetectable colorectal adenomas by sigmoidoscopy-
- Increased Prevalence of Colorectal Neoplasia in Korean Patients with Sporadic Duodenal Adenomas: A Case-Control Study