Clinical Application of Targeted Deep Sequencing in Metastatic Colorectal Cancer Patients: Actionable Genomic Alteration in K-MASTER Project
- Affiliations
-
- 1Department of Internal Medicine, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea
- 2K-MASTER Cancer Precision Medicine Diagnosis and Treatment Enterprise, Korea University Anam Hospital, Korea University College of Medicine, Seoul, Korea
- KMID: 2510654
- DOI: http://doi.org/10.4143/crt.2020.559
Abstract
- Purpose
Next-generation sequencing (NGS) can facilitate precision medicine approaches in metastatic colorectal cancer (mCRC) patients. We investigated the molecular profiling of Korean mCRC patients under the K-MASTER project which was initiated in June 2017 as a nationwide precision medicine oncology clinical trial platform which used NGS assay to screen actionable mutations.
Materials and Methods
As of 22 January 2020, total of 994 mCRC patients were registered in K-MASTER project. Targeted sequencing was performed using three platforms which were composed of the K-MASTER cancer panel v1.1 and the SNUH FIRST Cancer Panel v3.01. If tumor tissue was not available, cell-free DNA was extracted and the targeted sequencing was performed by Axen Cancer Panel as a liquid biopsy.
Results
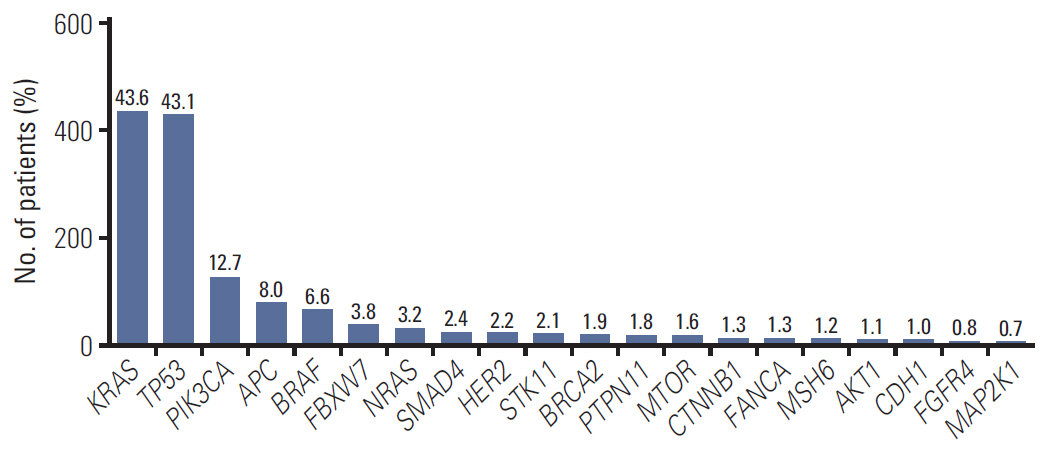
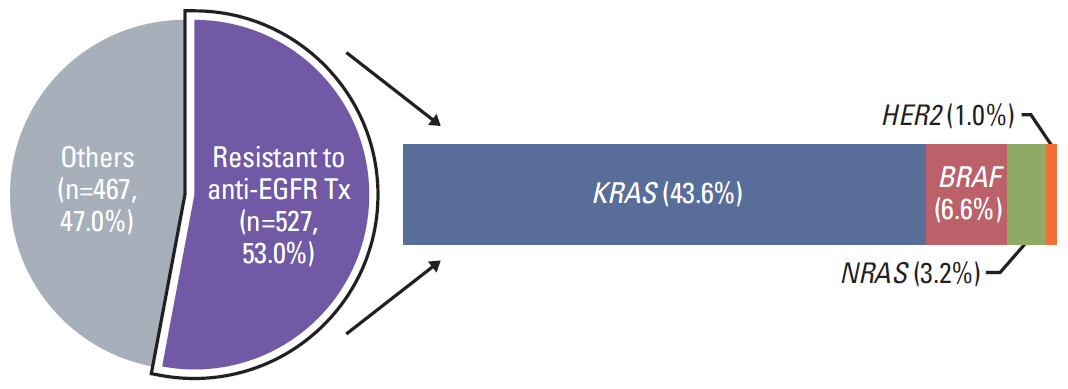
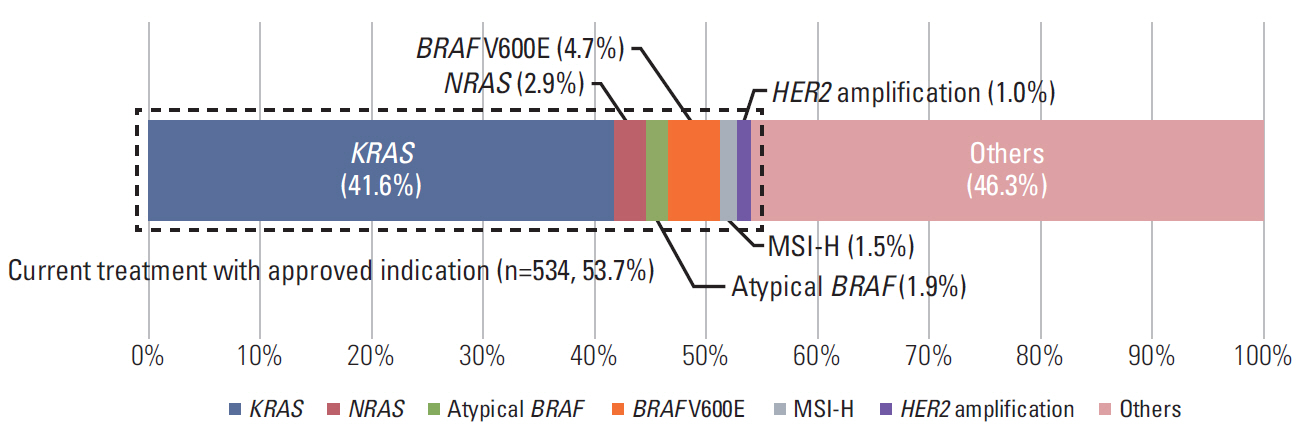
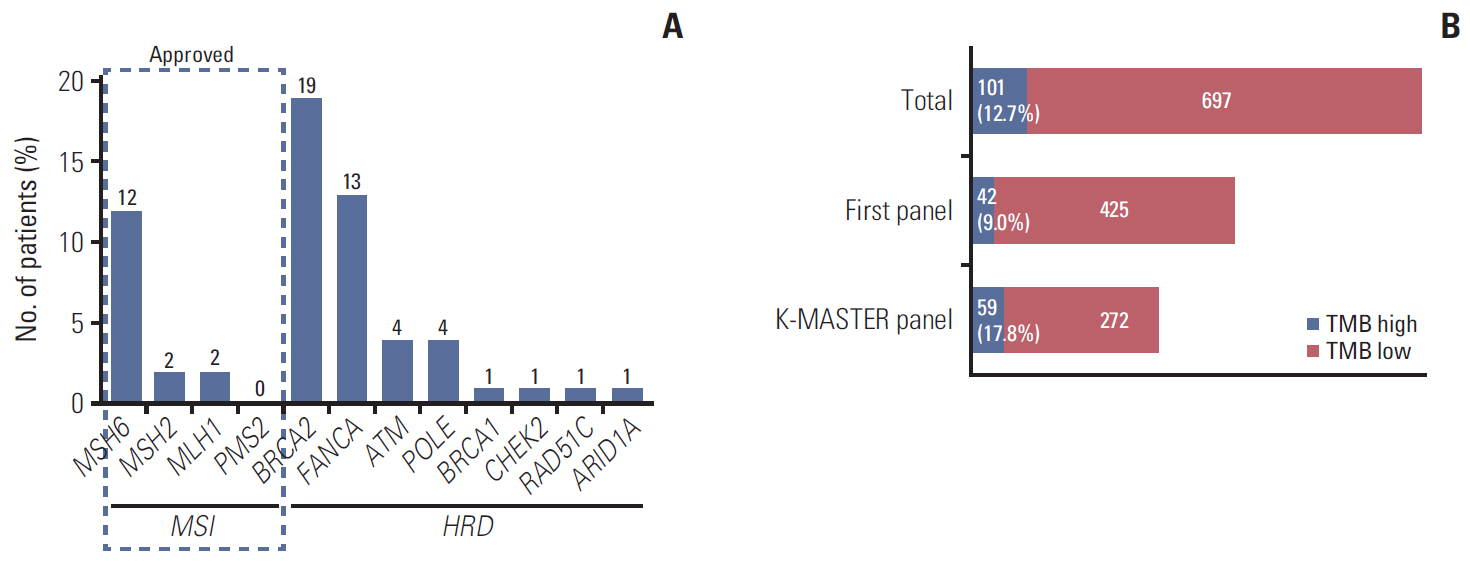
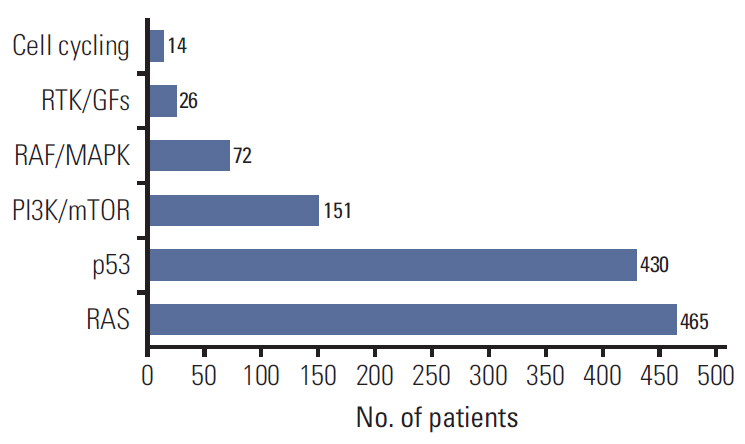
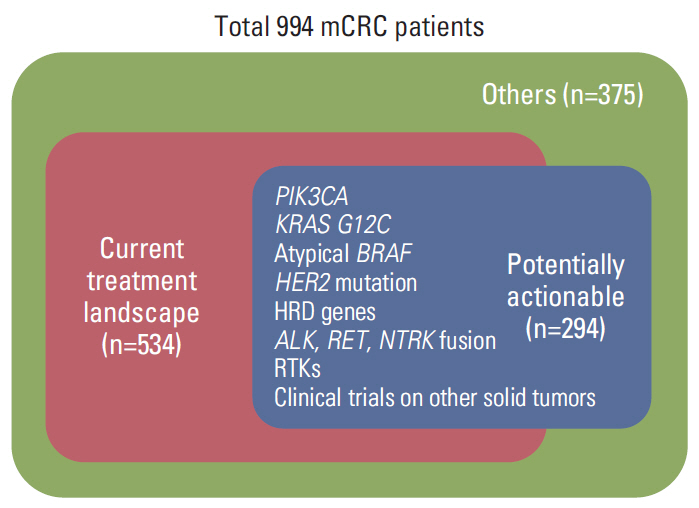
In 994 mCRC patients, we found 1,564 clinically meaningful pathogenic variants which mutated in 71 genes. Anti-EGFR therapy candidates were 467 patients (47.0%) and BRAF V600E mutation (n=47, 4.7%), deficient mismatch repair/microsatellite instability–high (n=15, 1.5%), HER2 amplifications (n=10, 1.0%) could be incorporated with recently approved drugs. The patients with high tumor mutation burden (n=101, 12.7%) and DNA damaging response and repair defect pathway alteration (n=42, 4.2%) could be enrolled clinical trials with immune checkpoint inhibitors. There were more colorectal cancer molecular alterations such as PIK3CA, KRAS G12C, atypical BRAF, and HER2 mutations and even rarer but actionable genes that approved or ongoing clinical trials in other solid tumors.
Conclusion
K-MASTER project provides an intriguing background to investigate new clinical trials with biomarkers and give therapeutic opportunity for mCRC patients.
Figure
Cited by 3 articles
-
Recommendations for the Use of Next-Generation Sequencing and the Molecular Tumor Board for Patients with Advanced Cancer: A Report from KSMO and KCSG Precision Medicine Networking Group
Shinkyo Yoon, Miso Kim, Yong Sang Hong, Han Sang Kim, Seung Tae Kim, Jihun Kim, Hongseok Yun, Changhoon Yoo, Hee Kyung Ahn, Hyo Song Kim, In Hee Lee, In-Ho Kim, Inkeun Park, Jae Ho Jeong, Jaekyung Cheon, Jin Won Kim, Jina Yun, Sun Min Lim, Yongjun Cha, Se Jin Jang, Dae Young Zang, Tae Won Kim, Jin Hyoung Kang, Jee Hyun Kim
Cancer Res Treat. 2022;54(1):1-9. doi: 10.4143/crt.2021.1115.Comparison of the Data of a Next-Generation Sequencing Panel from K-MASTER Project with That of Orthogonal Methods for Detecting Targetable Genetic Alterations
Yoon Ji Choi, Jung Yoon Choi, Ju Won Kim, Ah Reum Lim, Youngwoo Lee, Won Jin Chang, Soohyeon Lee, Jae Sook Sung, Hee-Joon Chung, Jong Won Lee, Eun Joo Kang, Jung Sun Kim, Taekyu Lim, Hye Sook Kim, Yu Jung Kim, Mi Sun Ahn, Young Saing Kim, Ji Hyun Park, Seungtaek Lim, Sung Shim Cho, Jang Ho Cho, Sang Won Shin, Kyong Hwa Park, Yeul Hong Kim
Cancer Res Treat. 2022;54(1):30-39. doi: 10.4143/crt.2021.218.PIK3CA Mutation Is Associated with Poor Response to HER2-Targeted Therapy in Breast Cancer Patients
Ju Won Kim, Ah Reum Lim, Ji Young You, Jung Hyun Lee, Sung Eun Song, Nam Kwon Lee, Seung Pil Jung, Kyu Ran Cho, Cheol Yong Kim, Kyong Hwa Park
Cancer Res Treat. 2023;55(2):531-541. doi: 10.4143/crt.2022.221.
Reference
-
References
1. Jung KW, Won YJ, Kong HJ, Lee ES. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res Treat. 2019; 51:417–30.
Article2. Choi JY, Choi S, Lee M, Park YS, Sung JS, Chang WJ, et al. Clinical implication of concordant or discordant genomic profiling between primary and matched metastatic tissues in patients with colorectal cancer. Cancer Res Treat. 2020; 52:764–78.
Article3. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.4. Shin HT, Choi YL, Yun JW, Kim NK, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017; 8:1377.
Article5. Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004; 351:337–45.
Article6. Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009; 360:1408–17.
Article7. Kim SY, Kim TW. Current challenges in the implementation of precision oncology for the management of metastatic colorectal cancer. ESMO Open. 2020; 5:e000634.
Article8. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019; 381:1632–43.9. Meric-Bernstam F, Hurwitz H, Raghav KP, McWilliams RR, Fakih M, VanderWalde A, et al. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019; 20:518–30.
Article10. Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016; 17:738–46.
Article11. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015; 372:2509–20.12. Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017; 18:1182–91.
Article13. Overman MJ, Lonardi S, Wong KY, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/mcrosatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018; 36:773–9.14. Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD. DNA damage and repair biomarkers of immunotherapy Response. Cancer Discov. 2017; 7:675–93.
Article15. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017; 9:34.
Article16. Lee DW, Han SW, Bae JM, Jang H, Han H, Kim H, et al. Tumor mutation burden and prognosis in patients with colorectal cancer treated with adjuvant fluoropyrimidine and oxaliplatin. Clin Cancer Res. 2019; 25:6141–7.
Article17. Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017; 109:djx089.
Article18. Gautschi O, Milia J, Filleron T, Wolf J, Carbone DP, Owen D, et al. Targeting RET in patients with RET-rearranged lung cancers: results from the global, multicenter RET registry. J Clin Oncol. 2017; 35:1403–10.19. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018; 378:731–9.20. Drilon A, Siena S, Ou SI, Patel M, Ahn MJ, Lee J, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: combined results from two phase I trials (ALKA-372-001 and STARTRK-1). Cancer Discov. 2017; 7:400–9.21. Dos Santos W, Sobanski T, de Carvalho AC, Evangelista AF, Matsushita M, Berardinelli GN, et al. Mutation profiling of cancer drivers in Brazilian colorectal cancer. Sci Rep. 2019; 9:13687.
Article22. D’Haene N, Fontanges Q, De Neve N, Blanchard O, Melendez B, Delos M, et al. Clinical application of targeted next-generation sequencing for colorectal cancer patients: a multicentric Belgian experience. Oncotarget. 2018; 9:20761–8.
Article23. Salem ME, Weinberg BA, Xiu J, El-Deiry WS, Hwang JJ, Gatalica Z, et al. Comparative molecular analyses of left-sided colon, right-sided colon, and rectal cancers. Oncotarget. 2017; 8:86356–68.
Article24. Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016; 17:1206.
Article25. Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, et al. International Cancer Genome Consortium Data Portal: a one-stop shop for cancer genomics data. Database (Oxford). 2011; 2011:bar026.26. Dong Z, Kong L, Wan Z, Zhu F, Zhong M, Lv Y, et al. Somatic mutation profiling and HER2 status in KRAS-positive Chinese colorectal cancer patients. Sci Rep. 2019; 9:16894.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Cancer Precision Medicine Diagnosis and Treatment (K-MASTER) Enterprise
- Genomic Profiling of Prostate Cancer: An Updated Review
- A Universal Analysis Pipeline for Hybrid Capture-Based Targeted Sequencing Data with Unique Molecular Indexes
- Update on Diagnosis and Treatment of Colorectal Cancer
- Comparison of the Data of a Next-Generation Sequencing Panel from K-MASTER Project with That of Orthogonal Methods for Detecting Targetable Genetic Alterations