J Vet Sci.
2020 Mar;21(2):e25. 10.4142/jvs.2020.21.e25.
A descriptive study of on-farm biosecurity and management practices during the incursion of porcine epidemic diarrhea into Canadian swine herds, 2014
- Affiliations
-
- 1Department of Population Medicine, Ontario Veterinary College, University of Guelph, Guelph, Ontario N1G 2W1, Canada
- 2Department of Large Animal Clinical Sciences, Western College of Veterinary Medicine, Saskatoon, Saskatchewan S7N 5A2, Canada
- KMID: 2506186
- DOI: http://doi.org/10.4142/jvs.2020.21.e25
Abstract
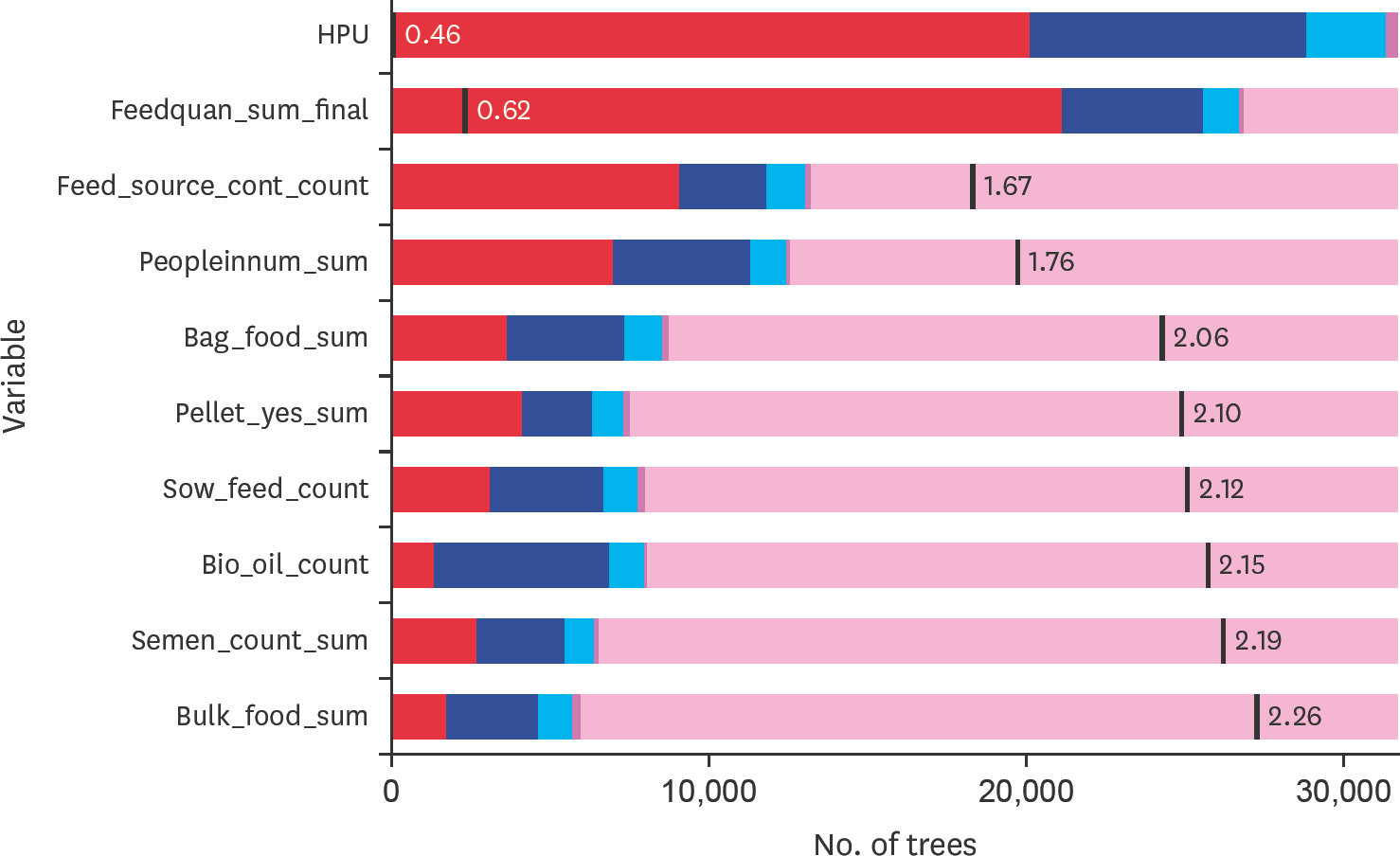
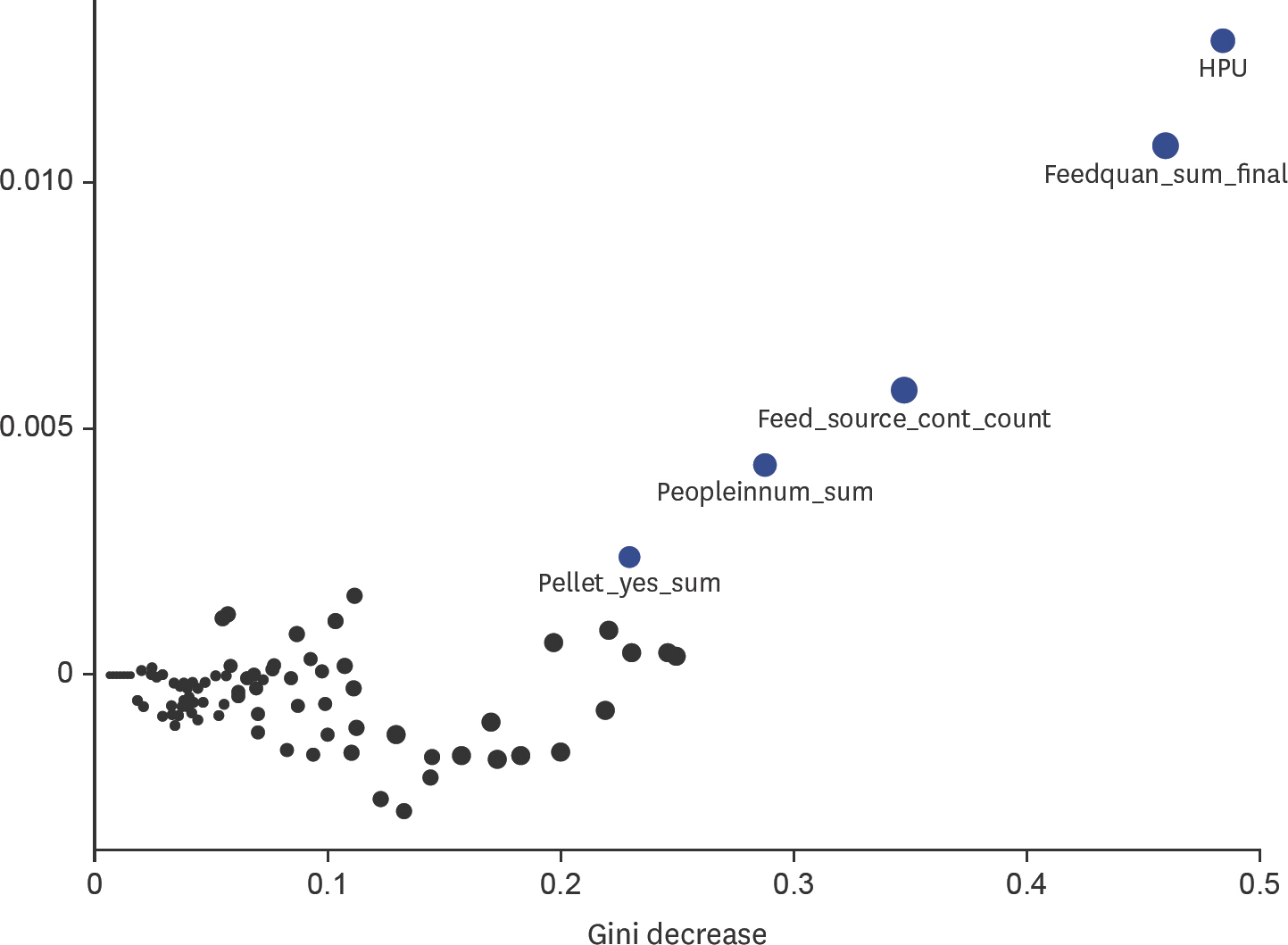
- Porcine epidemic diarrhea virus (PEDV) emerged into Canada in January 2014, primarily affecting sow herds. Subsequent epidemiological analyses suggested contaminated feed was the most likely transmission pathway. The primary objective of this study was to describe general biosecurity and management practices implemented in PEDV-positive sow herds and matched control herds at the time the virus emerged. The secondary objective was to determine if any of these general biosecurity and farm management practices were important in explaining PEDV infection status from January 22, 2014 to March 1, 2014. A case herd was defined as a swine herd with clinical signs and a positive test result for PEDV. A questionnaire was used to a gather 30-day history of herd management practices, animal movements on/off site, feed management practices, semen deliveries and biosecurity practices for case (n = 8) and control (n = 12) herds, primarily located in Ontario. Data was analyzed using descriptive statistics and random forests (RFs). Case herds were larger in size than control herds. Case herds had more animal movements and non-staff movements onto the site. Also, case herds had higher quantities of pigs delivered, feed deliveries and semen deliveries on-site. The biosecurity practices of case herds were considered more rigorous based on herd management, feed deliveries, transportation and truck driver practices than control herds. The RF model found that the most important variables for predicting herd status were related to herd size and feed management variables. Nonetheless, predictive accuracy of the final RF model was 72%.
Keyword
Figure
Reference
-
References
1. EFSA Panel on Animal Health and Welfare (AHAW). Scientific opinion on porcine epidemic diarrhoea and emerging porcine deltacoronavirus. EFSA J. 2014; 12:3877.2. Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013; 25:649–654.
Article3. Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014; 20:917–919.
Article4. Pasick J, Berhane Y, Ojkic D, Maxie G, Embury-Hyatt C, Swekla K, Handel K, Fairles J, Alexandersen S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound Emerg Dis. 2014; 61:397–410.
Article5. Kim YK, Lim SI, Cho IS, Cheong KM, Lee EJ, Lee SO, Kim JB, Kim JH, Jeong DS, An BH, An DJ. A novel diagnostic approach to detecting porcine epidemic diarrhea virus: The lateral immunochromatography assay. J Virol Methods. 2015; 225:4–8.
Article6. Alvarez J, Goede D, Morrison R, Perez A. Spatial and temporal epidemiology of porcine epidemic diarrhea (PED) in the Midwest and Southeast regions of the United States. Prev Vet Med. 2016; 123:155–160.
Article7. Ayudhya SN, Assavacheep P, Thanawongnuwech R, Thanawongnuwech R. One world–one health: the threat of emerging swine diseases. an Asian perspective. Transbound Emerg Dis. 2012; 59(Suppl 1):9–17.8. Pospischil A, Stuedli A, Kiupel M. Update on porcine epidemic diarrhea. J Swine Health Prod. 2002; 10:81–85.9. Sasaki Y, Alvarez J, Sekiguchi S, Sueyoshi M, Otake S, Perez A. Epidemiological factors associated to spread of porcine epidemic diarrhea in Japan. Prev Vet Med. 2016; 123:161–167.
Article10. Dee S, Neill C, Clement T, Singrey A, Christopher-Hennings J, Nelson E. An evaluation of porcine epidemic diarrhea virus survival in individual feed ingredients in the presence or absence of a liquid antimicrobial. Porcine Health Manag. 2015; 1:9.
Article11. Opriessnig T, Xiao CT, Gerber PF, Zhang J, Halbur PG, Halbur P. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naïve pigs. PLoS One. 2014; 9:e104766.
Article12. Perri AM, Poljak Z, Dewey C, Harding JCS, O'Sullivan TL. An epidemiological investigation of the early phase of the porcine epidemic diarrhea (PED) outbreak in Canadian swine herds in 2014: A case-control study. Prev Vet Med. 2018; 150:101–109.
Article13. Perri AM, Poljak Z, Dewey C, Harding JCS, O'Sullivan TL. Network analyses using case-control data to describe and characterize the initial 2014 incursion of porcine epidemic diarrhea (PED) in Canadian swine herds. Prev Vet Med. 2019; 162:18–28.
Article14. Pasma T, Furness MC, Alves D, Aubry P. Outbreak investigation of porcine epidemic diarrhea in swine in Ontario. Can Vet J. 2016; 57:84–89.15. Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012; 99:323–329.
Article16. Flori J, Mousing J, Gardner I, Willeberg P, Have P. Risk factors associated with seropositivity to porcine respiratory coronavirus in Danish swine herds. Prev Vet Med. 1995; 25:51–62.
Article17. Lambert ME, Arsenault J, Poljak Z, D'Allaire S. Epidemiological investigations in regard to porcine reproductive and respiratory syndrome (PRRS) in Quebec, Canada. Part 2: prevalence and risk factors in breeding sites. Prev Vet Med. 2012; 104:84–93.
Article18. Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002; 2:18–22.19. Kuhn M. Classification and Regression Training. [updated 2018;cited 2018 September 19]. Available from:. https://cran.r-project.org/web/packages/caret/caret.pdf.20. Paluszyńska A. Understanding Random Forests with randomForestExplainer. [updated 2017;cited 2018 September 19]. Available from:. https://cran.rstudio.com/web/packages/randomForestExplainer/vignettes/randomForestExplainer.html.21. Jung K, Saif LJ. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet J. 2015; 204:134–143.
Article22. United States Department of Agriculture Economics, Statistics and Market Information System. Overview of the United States Hog Industry. [updated 2015;cited 2018 October 1]. Available from:. http://usda.mannlib.cornell.edu/usda/current/hogview/hogview-10-29-2015.pdf.23. Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. The first case of porcine epidemic diarrhea in Canada. Can Vet J. 2015; 56:149–152.24. Lowe J, Gauger P, Harmon K, Zhang J, Connor J, Yeske P, Loula T, Levis I, Dufresne L, Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg Infect Dis. 2014; 20:872–874.
Article25. Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. Effects of air temperature and relative humidity on coronavirus survival on surfaces. Appl Environ Microbiol. 2010; 76:2712–2717.
Article26. Strobl C, Boulesteix AL, Zeileis A, Hothorn T. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics. 2007; 8:25.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Assessing the risk of recurrence of porcine epidemic diarrhea virus in affected farms on Jeju Island, South Korea
- Porcine epidemic diarrhea virus: an update overview of virus epidemiology, vaccines, and control strategies in South Korea
- Isolation and characterization of a new porcine epidemic diarrhea virus variant that occurred in Korea in 2014
- Porcine epidemic diarrhea: a review of current epidemiology and available vaccines
- Reemergence of porcine epidemic diarrhea virus on Jeju Island