Ann Surg Treat Res.
2020 Jul;99(1):44-51. 10.4174/astr.2020.99.1.44.
The effect of curative resection on fecal microbiota in patients with colorectal cancer: a prospective pilot study
- Affiliations
-
- 1Center for Colorectal Cancer, Research Institute and Hospital, National Cancer Center, Goyang, Korea
- KMID: 2503459
- DOI: http://doi.org/10.4174/astr.2020.99.1.44
Abstract
- Purpose
Although many studies have evaluated the association between intestinal microorganisms and the risk of colorectal cancer (CRC), only a few studies have investigated the changes in microorganisms following curative treatment for CRC. The current study analyzed changes in intestinal microbiota following curative surgery in CRC patients.
Methods
Stool samples were collected before and 6 months after surgery, from 11 patients with clinical stage III CRC, who underwent curative surgery between May 2017 and June 2017. Next, 16S rRNA gene sequencing was performed. Operational taxonomic units (OTUs) and alpha diversity were evaluated using the Shannon index. The bacterial compositions of the stools were analyzed according to taxonomic rank at genus and phylum levels.
Results
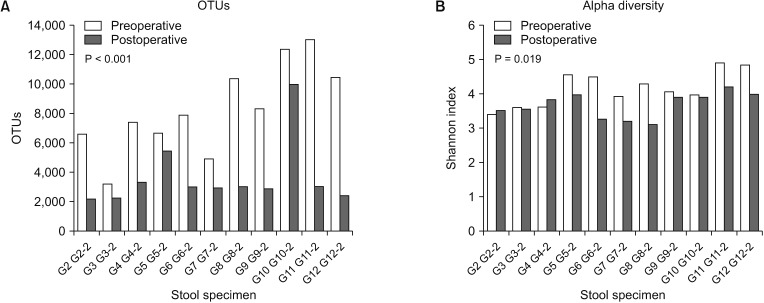
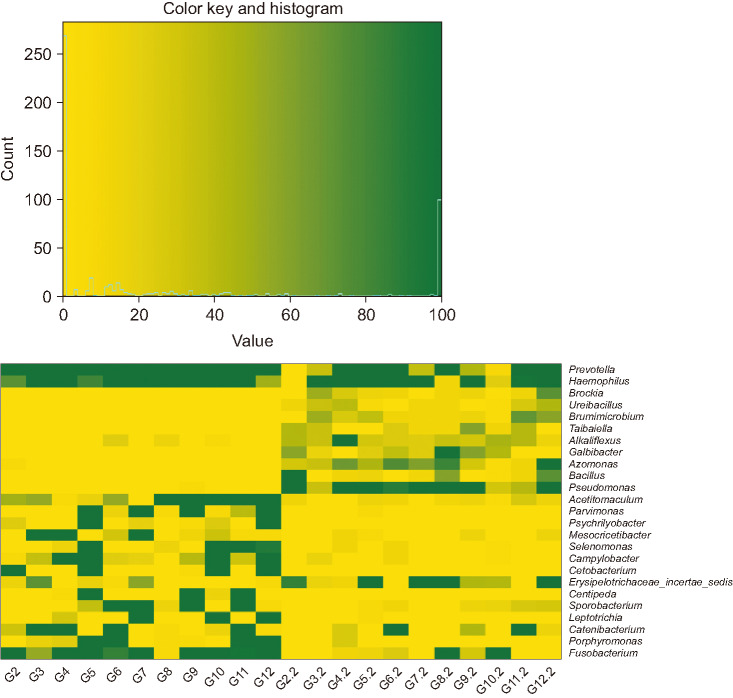
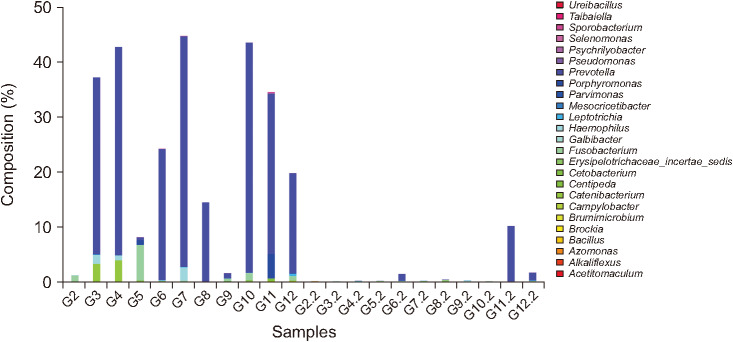
OTUs and alpha diversity were significantly decreased following surgery (P < 0.001 and P = 0.019, respectively). The compositions of several bacterial taxa changed after surgery. At genus level, proportions of pathogens such as Campylobacter, Fusobacterium, Haemophilus, Porphyromonas, and Prevotella, decreased after surgery (adjusted P < 0.05). At phylum level, the proportion of Fusobacteria decreased after surgery (adjusted P < 0.001).
Conclusion
Significant changes in intestinal microbial communities were noted following curative resection of CRC patients. Especially, decreases in pathogenic bacterial populations, such as Fusobacterium and Prevotella, which are known to be associated with CRC development, were detected even though OTUs and alpha diversity were decreased following curative resection. To determine and validate the clinical significance of these findings, large scale, prospective studies that include cancer prognoses are required.
Keyword
Figure
Cited by 1 articles
-
The effect of probiotics supplementation in postoperative cancer patients: a prospective pilot study
Hyeji Kwon, Song Hwa Chae, Hyo Jin Jung, Hyeon Min Shin, O-Hyun Ban, Jungwoo Yang, Jung Ha Kim, Ji Eun Jeong, Hae Myung Jeon, Yong Won Kang, Chan Kum Park, Daeyoun David Won, Jong Kyun Lee
Ann Surg Treat Res. 2021;101(5):281-290. doi: 10.4174/astr.2021.101.5.281.
Reference
-
1. Uhry Z, Belot A, Colonna M, Bossard N, Rogel A, Iwaz J, et al. National cancer incidence is estimated using the incidence/mortality ratio in countries with local incidence data: is this estimation correct? Cancer Epidemiol. 2013; 37:270–277. PMID: 23312453.2. Kim JC. Genopathogenesis of Colorectal Cancer. J Korean Surg Soc. 2005; 69:189–198.3. Berstad P, Loberg M, Larsen IK, Kalager M, Holme O, Botteri E, et al. Long-term lifestyle changes after colorectal cancer screening: randomised controlled trial. Gut. 2015; 64:1268–1276. PMID: 25183203.4. Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Version 2. Genes Dis. 2016; 3:130–143. PMID: 28078319.5. Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr. 2013; 97:1044–1052. PMID: 23553152.6. Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009; 15:1524–1527. PMID: 19442169.7. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012; 6:320–329. PMID: 21850056.8. Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013; 34:1285–1300. PMID: 23397545.9. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006; 124:837–848. PMID: 16497592.10. Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011; 11:9–20. PMID: 21151034.11. Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, et al. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016; 4:69. PMID: 28038683.12. Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012; 6:1858–1868. PMID: 22622349.13. Sze MA, Baxter NT, Ruffin MT 4th, Rogers MAM, Schloss PD. Normalization of the microbiota in patients after treatment for colonic lesions. Microbiome. 2017; 5:150. PMID: 29145893.14. Gallimore AM, Godkin A. Epithelial barriers, microbiota, and colorectal cancer. N Engl J Med. 2013; 368:282–284. PMID: 23323906.15. Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012; 491:254–258. PMID: 23034650.16. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015; 6:8727. PMID: 26515465.17. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015; 6:6528. PMID: 25758642.18. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27:2957–2963. PMID: 21903629.19. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7:335–336. PMID: 20383131.20. Flemer B, Lynch DB, Brown JM, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017; 66:633–643. PMID: 26992426.21. Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, et al. Comparison of human gut microbiota in control subjects and patients with colorectal carcinoma in adenoma: terminal restriction fragment length polymorphism and next-generation sequencing analyses. Oncol Rep. 2016; 35:325–333. PMID: 26549775.22. Sinha R, Ahn J, Sampson JN, Shi J, Yu G, Xiong X, et al. Fecal microbiota, fecal metabolome, and colorectal cancer interrelations. PLoS One. 2016; 11:e0152126. PMID: 27015276.23. Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016; 65:1973–1980. PMID: 26311717.24. Cong J, Zhu H, Liu D, Li T, Zhang C, Zhu J, et al. A pilot study: changes of gut microbiota in post-surgery colorectal cancer patients. Front Microbiol. 2018; 9:2777. PMID: 30515141.25. Jin Y, Liu Y, Zhao L, Zhao F, Feng J, Li S, et al. Gut microbiota in patients after surgical treatment for colorectal cancer. Environ Microbiol. 2019; 21:772–783. PMID: 30548192.26. Kong C, Gao R, Yan X, Huang L, He J, Li H, et al. Alterations in intestinal microbiota of colorectal cancer patients receiving radical surgery combined with adjuvant CapeOx therapy. Sci China Life Sci. 2019; 62:1178–1193. PMID: 30796721.27. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011; 108 Suppl 1(Suppl 1):4554–4561. PMID: 20847294.28. Zwielehner J, Lassl C, Hippe B, Pointner A, Switzeny OJ, Remely M, et al. Changes in human fecal microbiota due to chemotherapy analyzed by TaqMan-PCR, 454 sequencing and PCR-DGGE fingerprinting. PLoS One. 2011; 6:e28654. PMID: 22194876.29. Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014; 10:766. PMID: 25432777.30. Lin XH, Jiang JK, Luo JC, Lin CC, Ting PH, Yang UC, et al. The long term microbiota and metabolic status in patients with colorectal cancer after curative colon surgery. PLoS One. 2019; 14:e0218436. PMID: 31199857.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- CORRIGENDUM: The effect of curative resection on fecal microbiota in patients with colorectal cancer: a prospective pilot study
- Fecal Microbiota Transplantation and the Brain Microbiota in Neurological Diseases
- Effect of preoperative immunonutrition on fecal microbiota in colon cancer patients: a secondary analysis of a randomized controlled trial
- Gut Microbiome and Colorectal Cancer
- Commensal Microbiota and Cancer Immunotherapy: Harnessing Commensal Bacteria for Cancer Therapy