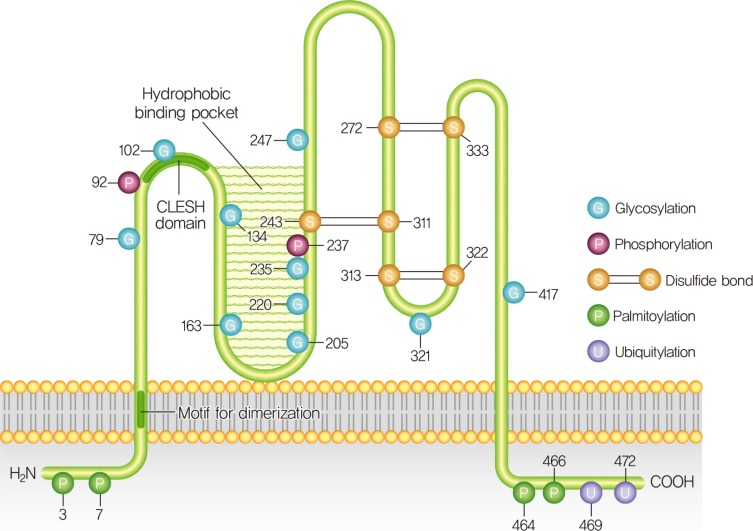
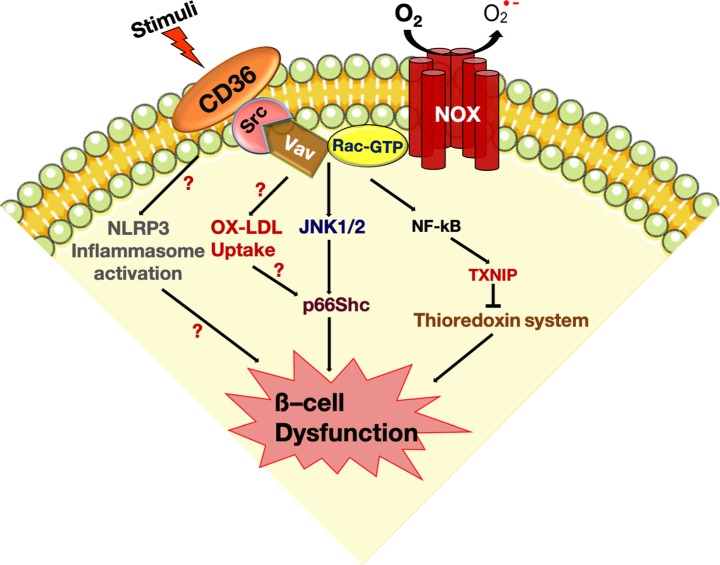
Diabetes Metab J.
2020 Apr;44(2):222-233. 10.4093/dmj.2020.0053.
The Role of CD36 in Type 2 Diabetes Mellitus: β-Cell Dysfunction and Beyond
- Affiliations
-
- 1Department of Internal Medicine, Yeungnam University College of Medicine, Daegu, Korea
- KMID: 2502403
- DOI: http://doi.org/10.4093/dmj.2020.0053
Abstract
- Impaired β-cell function is the key pathophysiology of type 2 diabetes mellitus, and chronic exposure of nutrient excess could lead to this tragedy. For preserving β-cell function, it is essential to understand the cause and mechanisms about the progression of β-cells failure. Glucotoxicity, lipotoxicity, and glucolipotoxicity have been suggested to be a major cause of β-cell dysfunction for decades, but not yet fully understood. Fatty acid translocase cluster determinant 36 (CD36), which is part of the free fatty acid (FFA) transporter system, has been identified in several tissues such as muscle, liver, and insulin-producing cells. Several studies have reported that induction of CD36 increases uptake of FFA in several cells, suggesting the functional interplay between glucose and FFA in terms of insulin secretion and oxidative metabolism. However, we do not currently know the regulating mechanism and physiological role of CD36 on glucolipotoxicity in pancreatic β-cells. Also, the downstream and upstream targets of CD36 related signaling have not been defined. In the present review, we will focus on the expression and function of CD36 related signaling in the pancreatic β-cells in response to hyperglycemia and hyperlipidemia (ceramide) along with the clinical studies on the association between CD36 and metabolic disorders.
Keyword
Figure
Reference
-
1. Kim BY, Won JC, Lee JH, Kim HS, Park JH, Ha KH, Won KC, Kim DJ, Park KS. Diabetes fact sheets in Korea, 2018: an appraisal of current status. Diabetes Metab J. 2019; 43:487–494. PMID: 31339012.
Article2. Schwartz SS, Epstein S, Corkey BE, Grant SF, Gavin JR 3rd, Aguilar RB. The time is right for a new classification system for diabetes: rationale and implications of the β-cell-centric classification schema. Diabetes Care. 2016; 39:179–186. PMID: 26798148.
Article3. Abumrad NA, el-Maghrabi MR, Amri EZ, Lopez E, Grimaldi PA. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation: homology with human CD36. J Biol Chem. 1993; 268:17665–17668. PMID: 7688729.
Article4. Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem. 1994; 269:18985–18991. PMID: 7518447.
Article5. Puente Navazo MD, Daviet L, Ninio E, McGregor JL. Identification on human CD36 of a domain (155-183) implicated in binding oxidized low-density lipoproteins (Ox-LDL). Arterioscler Thromb Vasc Biol. 1996; 16:1033–1039. PMID: 8696943.
Article6. Ohgami N, Nagai R, Ikemoto M, Arai H, Kuniyasu A, Horiuchi S, Nakayama H. CD36, a member of class B scavenger receptor family, is a receptor for advanced glycation end products. Ann N Y Acad Sci. 2001; 947:350–355. PMID: 11795289.
Article7. Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, Peters J, Neculai M, Plumb J, Loppnau P, Pizarro JC, Seitova A, Trimble WS, Saftig P, Grinstein S, Dhe-Paganon S. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013; 504:172–176. PMID: 24162852.
Article8. Wang J, Hao JW, Wang X, Guo H, Sun HH, Lai XY, Liu LY, Zhu M, Wang HY, Li YF, Yu LY, Xie C, Wang HR, Mo W, Zhou HM, Chen S, Liang G, Zhao TJ. DHHC4 and DHHC5 facilitate fatty acid uptake by palmitoylating and targeting CD36 to the plasma membrane. Cell Rep. 2019; 26:209–221. PMID: 30605677.
Article9. Zhao L, Zhang C, Luo X, Wang P, Zhou W, Zhong S, Xie Y, Jiang Y, Yang P, Tang R, Pan Q, Hall AR, Luong TV, Fan J, Varghese Z, Moorhead JF, Pinzani M, Chen Y, Ruan XZ. CD36 palmitoylation disrupts free fatty acid metabolism and promotes tissue inflammation in non-alcoholic steatohepatitis. J Hepatol. 2018; 69:705–717. PMID: 29705240.
Article10. Thorne RF, Ralston KJ, de Bock CE, Mhaidat NM, Zhang XD, Boyd AW, Burns GF. Palmitoylation of CD36/FAT regulates the rate of its post-transcriptional processing in the endoplasmic reticulum. Biochim Biophys Acta. 2010; 1803:1298–1307. PMID: 20637247.
Article11. Hoosdally SJ, Andress EJ, Wooding C, Martin CA, Linton KJ. The human scavenger receptor CD36: glycosylation status and its role in trafficking and function. J Biol Chem. 2009; 284:16277–16288. PMID: 19369259.12. Laczy B, Fulop N, Onay-Besikci A, Des Rosiers C, Chatham JC. Acute regulation of cardiac metabolism by the hexosamine biosynthesis pathway and protein O-GlcNAcylation. PLoS One. 2011; 6:e18417. PMID: 21494549.
Article13. Nabeebaccus AA, Zoccarato A, Hafstad AD, Santos CX, Aasum E, Brewer AC, Zhang M, Beretta M, Yin X, West JA, Schröder K, Griffin JL, Eykyn TR, Abel ED, Mayr M, Shah AM. Nox4 reprograms cardiac substrate metabolism via protein O-GlcNAcylation to enhance stress adaptation. JCI Insight. 2017; 2:96184. PMID: 29263294.
Article14. Asch AS, Liu I, Briccetti FM, Barnwell JW, Kwakye-Berko F, Dokun A, Goldberger J, Pernambuco M. Analysis of CD36 binding domains: ligand specificity controlled by dephosphorylation of an ectodomain. Science. 1993; 262:1436–1440. PMID: 7504322.
Article15. Hatmi M, Gavaret JM, Elalamy I, Vargaftig BB, Jacquemin C. Evidence for cAMP-dependent platelet ectoprotein kinase activity that phosphorylates platelet glycoprotein IV (CD36). J Biol Chem. 1996; 271:24776–24780. PMID: 8798748.
Article16. Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, Olsen JV. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012; 2:419–431. PMID: 22902405.
Article17. Smith J, Su X, El-Maghrabi R, Stahl PD, Abumrad NA. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J Biol Chem. 2008; 283:13578–13585. PMID: 18353783.18. Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL, Suen DF, Youle RJ, Amar M, Remaley AT, Sack MN. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011; 121:3701–3712. PMID: 21865652.
Article19. Noushmehr H, D'Amico E, Farilla L, Hui H, Wawrowsky KA, Mlynarski W, Doria A, Abumrad NA, Perfetti R. Fatty acid translocase (FAT/CD36) is localized on insulin-containing granules in human pancreatic beta-cells and mediates fatty acid effects on insulin secretion. Diabetes. 2005; 54:472–481. PMID: 15677505.20. Maedler K, Oberholzer J, Bucher P, Spinas GA, Donath MY. Monounsaturated fatty acids prevent the deleterious effects of palmitate and high glucose on human pancreatic beta-cell turnover and function. Diabetes. 2003; 52:726–733. PMID: 12606514.21. Maedler K, Spinas GA, Dyntar D, Moritz W, Kaiser N, Donath MY. Distinct effects of saturated and monounsaturated fatty acids on beta-cell turnover and function. Diabetes. 2001; 50:69–76. PMID: 11147797.22. Kim YW, Moon JS, Seo YJ, Park SY, Kim JY, Yoon JS, Lee IK, Lee HW, Won KC. Inhibition of fatty acid translocase cluster determinant 36 (CD36), stimulated by hyperglycemia, prevents glucotoxicity in INS-1 cells. Biochem Biophys Res Commun. 2012; 420:462–466. PMID: 22430143.
Article23. Wallin T, Ma Z, Ogata H, Jorgensen IH, Iezzi M, Wang H, Wollheim CB, Bjorklund A. Facilitation of fatty acid uptake by CD36 in insulin-producing cells reduces fatty-acid-induced insulin secretion and glucose regulation of fatty acid oxidation. Biochim Biophys Acta. 2010; 1801:191–197. PMID: 19931418.24. Glatz JF, Luiken JJ, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev. 2010; 90:367–417. PMID: 20086080.
Article25. Elumalai S, Karunakaran U, Lee IK, Moon JS, Won KC. Rac1-NADPH oxidase signaling promotes CD36 activation under glucotoxic conditions in pancreatic beta cells. Redox Biol. 2017; 11:126–134. PMID: 27912197.
Article26. Ihara Y, Toyokuni S, Uchida K, Odaka H, Tanaka T, Ikeda H, Hiai H, Seino Y, Yamada Y. Hyperglycemia causes oxidative stress in pancreatic beta-cells of GK rats, a model of type 2 diabetes. Diabetes. 1999; 48:927–932. PMID: 10102716.
Article27. Cnop M, Welsh N, Jonas JC, Jorns A, Lenzen S, Eizirik DL. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005; 54 Suppl 2:S97–S107. PMID: 16306347.28. Karunakaran U, Park KG. A systematic review of oxidative stress and safety of antioxidants in diabetes: focus on islets and their defense. Diabetes Metab J. 2013; 37:106–112. PMID: 23641350.
Article29. Guichard C, Moreau R, Pessayre D, Epperson TK, Krause KH. NOX family NADPH oxidases in liver and in pancreatic islets: a role in the metabolic syndrome and diabetes? Biochem Soc Trans. 2008; 36(Pt 5):920–929. PMID: 18793162.
Article30. Gharib M, Tao H, Fungwe TV, Hajri T. Cluster differentiating 36 (CD36) deficiency attenuates obesity-associated oxidative stress in the heart. PLoS One. 2016; 11:e0155611. PMID: 27195707.
Article31. Li W, Febbraio M, Reddy SP, Yu DY, Yamamoto M, Silverstein RL. CD36 participates in a signaling pathway that regulates ROS formation in murine VSMCs. J Clin Invest. 2010; 120:3996–4006. PMID: 20978343.
Article32. Jimenez B, Volpert OV, Crawford SE, Febbraio M, Silverstein RL, Bouck N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat Med. 2000; 6:41–48. PMID: 10613822.
Article33. Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW. A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J Biol Chem. 2002; 277:47373–47379. PMID: 12239221.34. Rahaman SO, Lennon DJ, Febbraio M, Podrez EA, Hazen SL, Silverstein RL. A CD36-dependent signaling cascade is necessary for macrophage foam cell formation. Cell Metab. 2006; 4:211–221. PMID: 16950138.
Article35. Lenzen S, Drinkgern J, Tiedge M. Low antioxidant enzyme gene expression in pancreatic islets compared with various other mouse tissues. Free Radic Biol Med. 1996; 20:463–466. PMID: 8720919.
Article36. Tiedge M, Lortz S, Drinkgern J, Lenzen S. Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes. 1997; 46:1733–1742. PMID: 9356019.
Article37. Haber EP, Ximenes HM, Procopio J, Carvalho CR, Curi R, Carpinelli AR. Pleiotropic effects of fatty acids on pancreatic beta-cells. J Cell Physiol. 2003; 194:1–12. PMID: 12447984.38. Prentki M, Nolan CJ. Islet beta cell failure in type 2 diabetes. J Clin Invest. 2006; 116:1802–1812. PMID: 16823478.39. Unger RH, Zhou YT. Lipotoxicity of beta-cells in obesity and in other causes of fatty acid spillover. Diabetes. 2001; 50 Suppl 1:S118–S121. PMID: 11272168.
Article40. Briaud I, Harmon JS, Kelpe CL, Segu VB, Poitout V. Lipotoxicity of the pancreatic beta-cell is associated with glucose-dependent esterification of fatty acids into neutral lipids. Diabetes. 2001; 50:315–321. PMID: 11272142.41. Lupi R, Dotta F, Marselli L, Del Guerra S, Masini M, Santangelo C, Patane G, Boggi U, Piro S, Anello M, Bergamini E, Mosca F, Di Mario U, Del Prato S, Marchetti P. Prolonged exposure to free fatty acids has cytostatic and pro-apoptotic effects on human pancreatic islets: evidence that beta-cell death is caspase mediated, partially dependent on ceramide pathway, and Bcl-2 regulated. Diabetes. 2002; 51:1437–1442. PMID: 11978640.42. Shimabukuro M, Ohneda M, Lee Y, Unger RH. Role of nitric oxide in obesity-induced beta cell disease. J Clin Invest. 1997; 100:290–295. PMID: 9218505.
Article43. Maestre I, Jordan J, Calvo S, Reig JA, Cena V, Soria B, Prentki M, Roche E. Mitochondrial dysfunction is involved in apoptosis induced by serum withdrawal and fatty acids in the beta-cell line INS-1. Endocrinology. 2003; 144:335–345. PMID: 12488362.44. Chen M, Yang YK, Loux TJ, Georgeson KE, Harmon CM. The role of hyperglycemia in FAT/CD36 expression and function. Pediatr Surg Int. 2006; 22:647–654. PMID: 16838191.
Article45. Farhangkhoee H, Khan ZA, Barbin Y, Chakrabarti S. Glucose-induced up-regulation of CD36 mediates oxidative stress and microvascular endothelial cell dysfunction. Diabetologia. 2005; 48:1401–1410. PMID: 15915335.
Article46. Xue JH, Yuan Z, Wu Y, Liu Y, Zhao Y, Zhang WP, Tian YL, Liu WM, Liu Y, Kishimoto C. High glucose promotes intracellular lipid accumulation in vascular smooth muscle cells by impairing cholesterol influx and efflux balance. Cardiovasc Res. 2010; 86:141–150. PMID: 20007688.
Article47. Shimabukuro M, Zhou YT, Levi M, Unger RH. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc Natl Acad Sci U S A. 1998; 95:2498–2502. PMID: 9482914.48. Samad F, Hester KD, Yang G, Hannun YA, Bielawski J. Altered adipose and plasma sphingolipid metabolism in obesity: a potential mechanism for cardiovascular and metabolic risk. Diabetes. 2006; 55:2579–2587. PMID: 16936207.49. Haus JM, Kashyap SR, Kasumov T, Zhang R, Kelly KR, Defronzo RA, Kirwan JP. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 2009; 58:337–343. PMID: 19008343.
Article50. Wigger L, Cruciani-Guglielmacci C, Nicolas A, Denom J, Fernandez N, Fumeron F, Marques-Vidal P, Ktorza A, Kramer W, Schulte A, Le Stunff H, Liechti R, Xenarios I, Vollenweider P, Waeber G, Uphues I, Roussel R, Magnan C, Ibberson M, Thorens B. Plasma dihydroceramides are diabetes susceptibility biomarker candidates in mice and humans. Cell Rep. 2017; 18:2269–2279. PMID: 28249170.
Article51. Bustelo XR. Vav family exchange factors: an integrated regulatory and functional view. Small GTPases. 2014; 5:9. PMID: 25483299.
Article52. Kominato R, Fujimoto S, Mukai E, Nakamura Y, Nabe K, Shimodahira M, Nishi Y, Funakoshi S, Seino Y, Inagaki N. Src activation generates reactive oxygen species and impairs metabolism-secretion coupling in diabetic Goto-Kakizaki and ouabain-treated rat pancreatic islets. Diabetologia. 2008; 51:1226–1235. PMID: 18449527.
Article53. Karunakaran U, Elumalai S, Moon JS, Won KC. CD36 dependent redoxosomes promotes ceramide-mediated pancreatic β-cell failure via p66Shc activation. Free Radic Biol Med. 2019; 134:505–515. PMID: 30735834.
Article54. Holzer RG, Park EJ, Li N, Tran H, Chen M, Choi C, Solinas G, Karin M. Saturated fatty acids induce c-Src clustering within membrane subdomains, leading to JNK activation. Cell. 2011; 147:173–184. PMID: 21962514.
Article55. Kant S, Standen CL, Morel C, Jung DY, Kim JK, Swat W, Flavell RA, Davis RJ. A protein scaffold coordinates SRC-mediated JNK activation in response to metabolic stress. Cell Rep. 2017; 20:2775–2783. PMID: 28930674.
Article56. Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002; 420:333–336. PMID: 12447443.
Article57. Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999; 402:309–313. PMID: 10580504.
Article58. Natalicchio A, Tortosa F, Labarbuta R, Biondi G, Marrano N, Carchia E, Leonardini A, Cignarelli A, Bugliani M, Marchetti P, Fadini GP, Giorgio M, Avogaro A, Perrini S, Laviola L, Giorgino F. The p66(Shc) redox adaptor protein is induced by saturated fatty acids and mediates lipotoxicity-induced apoptosis in pancreatic beta cells. Diabetologia. 2015; 58:1260–1271. PMID: 25810038.
Article59. Khalid S, Drasche A, Thurner M, Hermann M, Ashraf MI, Fresser F, Baier G, Kremser L, Lindner H, Troppmair J. cJun N-terminal kinase (JNK) phosphorylation of serine 36 is critical for p66Shc activation. Sci Rep. 2016; 6:20930. PMID: 26868434.
Article60. Cox AG, Pullar JM, Hughes G, Ledgerwood EC, Hampton MB. Oxidation of mitochondrial peroxiredoxin 3 during the initiation of receptor-mediated apoptosis. Free Radic Biol Med. 2008; 44:1001–1009. PMID: 18164270.
Article61. Karunakaran U, Moon JS, Lee HW, Won KC. CD36 initiated signaling mediates ceramide-induced TXNIP expression in pancreatic beta-cells. Biochim Biophys Acta. 2015; 1852:2414–2422. PMID: 26297980.
Article62. Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001; 108:785–791. PMID: 11560944.
Article63. Grupping AY, Cnop M, Van Schravendijk CF, Hannaert JC, Van Berkel TJ, Pipeleers DG. Low density lipoprotein binding and uptake by human and rat islet beta cells. Endocrinology. 1997; 138:4064–4068. PMID: 9322913.64. Cnop M, Hannaert JC, Grupping AY, Pipeleers DG. Low density lipoprotein can cause death of islet beta-cells by its cellular uptake and oxidative modification. Endocrinology. 2002; 143:3449–3453. PMID: 12193557.65. Abderrahmani A, Niederhauser G, Favre D, Abdelli S, Ferdaoussi M, Yang JY, Regazzi R, Widmann C, Waeber G. Human high-density lipoprotein particles prevent activation of the JNK pathway induced by human oxidised low-density lipoprotein particles in pancreatic beta cells. Diabetologia. 2007; 50:1304–1314. PMID: 17437081.
Article66. Plaisance V, Brajkovic S, Tenenbaum M, Favre D, Ezanno H, Bonnefond A, Bonner C, Gmyr V, Kerr-Conte J, Gauthier BR, Widmann C, Waeber G, Pattou F, Froguel P, Abderrahmani A. Endoplasmic reticulum stress links oxidative stress to impaired pancreatic beta-cell function caused by human oxidized LDL. PLoS One. 2016; 11:e0163046. PMID: 27636901.
Article67. Moon JS, Karunakaran U, Elumalai S, Lee IK, Lee HW, Kim YW, Won KC. Metformin prevents glucotoxicity by alleviating oxidative and ER stress-induced CD36 expression in pancreatic beta cells. J Diabetes Complications. 2017; 31:21–30. PMID: 27662780.
Article68. Shi Y, Cosentino F, Camici GG, Akhmedov A, Vanhoutte PM, Tanner FC, Luscher TF. Oxidized low-density lipoprotein activates p66Shc via lectin-like oxidized low-density lipoprotein receptor-1, protein kinase C-beta, and c-Jun N-terminal kinase kinase in human endothelial cells. Arterioscler Thromb Vasc Biol. 2011; 31:2090–2097. PMID: 21817106.69. Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011; 17:179–188. PMID: 21217695.
Article70. Oslowski CM, Hara T, O'Sullivan-Murphy B, Kanekura K, Lu S, Hara M, Ishigaki S, Zhu LJ, Hayashi E, Hui ST, Greiner D, Kaufman RJ, Bortell R, Urano F. Thioredoxin-interacting protein mediates ER stress-induced β cell death through initiation of the inflammasome. Cell Metab. 2012; 16:265–273. PMID: 22883234.
Article71. Sheedy FJ, Grebe A, Rayner KJ, Kalantari P, Ramkhelawon B, Carpenter SB, Becker CE, Ediriweera HN, Mullick AE, Golenbock DT, Stuart LM, Latz E, Fitzgerald KA, Moore KJ. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol. 2013; 14:812–820. PMID: 23812099.
Article72. Koonen DP, Jensen MK, Handberg A. Soluble CD36- a marker of the (pathophysiological) role of CD36 in the metabolic syndrome? Arch Physiol Biochem. 2011; 117:57–63. PMID: 21250778.
Article73. Handberg A, Hojlund K, Gastaldelli A, Flyvbjerg A, Dekker JM, Petrie J, Piatti P, Beck-Nielsen H. RISC Investigators. Plasma sCD36 is associated with markers of atherosclerosis, insulin resistance and fatty liver in a nondiabetic healthy population. J Intern Med. 2012; 271:294–304. PMID: 21883535.
Article74. Handberg A, Levin K, Hojlund K, Beck-Nielsen H. Identification of the oxidized low-density lipoprotein scavenger receptor CD36 in plasma: a novel marker of insulin resistance. Circulation. 2006; 114:1169–1176. PMID: 16952981.75. Glintborg D, Hojlund K, Andersen M, Henriksen JE, Beck-Nielsen H, Handberg A. Soluble CD36 and risk markers of insulin resistance and atherosclerosis are elevated in polycystic ovary syndrome and significantly reduced during pioglitazone treatment. Diabetes Care. 2008; 31:328–334. PMID: 18000176.
Article76. Wang Y, Koch M, di Giuseppe R, Evans K, Borggrefe J, Nothlings U, Handberg A, Jensen MK, Lieb W. Associations of plasma CD36 and body fat distribution. J Clin Endocrinol Metab. 2019; jc.2019-00368.
Article77. Kim HJ, Moon JS, Park IR, Kim JH, Yoon JS, Won KC, Lee HW. A novel index using soluble CD36 is associated with the prevalence of type 2 diabetes mellitus: comparison study with triglyceride-glucose index. Endocrinol Metab (Seoul). 2017; 32:375–382. PMID: 28956368.
Article78. Hjuler Nielsen M, Irvine H, Vedel S, Raungaard B, Beck-Nielsen H, Handberg A. Elevated atherosclerosis-related gene expression, monocyte activation and microparticle-release are related to increased lipoprotein-associated oxidative stress in familial hypercholesterolemia. PLoS One. 2015; 10:e0121516. PMID: 25875611.
Article79. Pardina E, Ferrer R, Rossell J, Ricart-Jane D, Mendez-Lara KA, Baena-Fustegueras JA, Lecube A, Julve J, Peinado-Onsurbe J. Hepatic CD36 downregulation parallels steatosis improvement in morbidly obese undergoing bariatric surgery. Int J Obes (Lond). 2017; 41:1388–1393. PMID: 28555086.
Article80. Botha J, Nielsen MH, Christensen MH, Vestergaard H, Handberg A. Bariatric surgery reduces CD36-bearing microvesicles of endothelial and monocyte origin. Nutr Metab (Lond). 2018; 15:76. PMID: 30386406.
Article81. Al Dubayee MS, Alayed H, Almansour R, Alqaoud N, Alnamlah R, Obeid D, Alshahrani A, Zahra MM, Nasr A, Al-Bawab A, Aljada A. Differential expression of human peripheral mononuclear cells phenotype markers in type 2 diabetic patients and type 2 diabetic patients on metformin. Front Endocrinol (Lausanne). 2018; 9:537. PMID: 30356719.
Article82. Shiju TM, Mohan V, Balasubramanyam M, Viswanathan P. Soluble CD36 in plasma and urine: a plausible prognostic marker for diabetic nephropathy. J Diabetes Complications. 2015; 29:400–406. PMID: 25619588.
Article83. Castelblanco E, Sanjurjo L, Falguera M, Hernandez M, Fernandez-Real JM, Sarrias MR, Alonso N, Mauricio D. Circulating soluble CD36 is similar in type 1 and type 2 diabetes mellitus versus non-diabetic subjects. J Clin Med. 2019; 8:E710. PMID: 31109109.
Article84. Wang Y, Zhu J, Aroner S, Overvad K, Cai T, Yang M, Tjonneland A, Handberg A, Jensen MK. Plasma CD36 and incident diabetes: a case-cohort study in Danish men and women. Diabetes Metab J. 2020; 44:134–142. PMID: 31701685.
Article85. Jiang X, Zhao X, Chen R, Jiang Q, Zhou B. Plasma soluble CD36, carotid intima-media thickness and cognitive function in patients with type 2 diabetes. Arch Med Sci. 2017; 13:1031–1039. PMID: 28883843.
Article86. Handberg A, Skjelland M, Michelsen AE, Sagen EL, Krohg-Sorensen K, Russell D, Dahl A, Ueland T, Oie E, Aukrust P, Halvorsen B. Soluble CD36 in plasma is increased in patients with symptomatic atherosclerotic carotid plaques and is related to plaque instability. Stroke. 2008; 39:3092–3095. PMID: 18723424.
Article87. Wang Y, Zhu J, Handberg A, Overvad K, Tjønneland A, Rimm EB, Jensen MK. Association between plasma CD36 levels and incident risk of coronary heart disease among Danish men and women. Atherosclerosis. 2018; 277:163–168. PMID: 30218892.
Article88. Gautam S, Pirabu L, Agrawal CG, Banerjee M. CD36 gene variants and their association with type 2 diabetes in an Indian population. Diabetes Technol Ther. 2013; 15:680–687. PMID: 23844572.
Article89. Zhang Y, Zang J, Wang B, Li B, Yao X, Zhao H, Li W. CD36 genotype associated with ischemic stroke in Chinese Han. Int J Clin Exp Med. 2015; 8:16149–16157. PMID: 26629128.90. Zhang D, Zhang R, Liu Y, Sun X, Yin Z, Li H, Zhao Y, Wang B, Ren Y, Cheng C, Liu X, Liu D, Liu F, Chen X, Liu L, Zhou Q, Xiong Y, Xu Q, Liu J, Hong S, You Z, Hu D, Zhang M. CD36 gene variants is associated with type 2 diabetes mellitus through the interaction of obesity in rural Chinese adults. Gene. 2018; 659:155–159. PMID: 29572193.
Article