J Pathol Transl Med.
2020 Jan;54(1):45-63. 10.4132/jptm.2019.11.05.
Molecular characteristics of meningiomas
- Affiliations
-
- 1Department of Hospital Pathology, College of Medicine, The Catholic University of Korea, Seoul, Korea
- KMID: 2501594
- DOI: http://doi.org/10.4132/jptm.2019.11.05
Abstract
- Meningioma is the most common primary intracranial tumor in adults. The grading of meningioma is based on World Health Organization criteria, which rely on histopathological features alone. This grading system is unable to conclusively predict the clinical behavior of these tumors (i.e., recurrence or prognosis in benign or atypical grades). Advances in molecular techniques over the last decade that include genomic and epigenomic data associated with meningiomas have been used to identify genetic biomarkers that can predict tumor behavior. This review summarizes the molecular characteristics of meningioma using genetic and epigenetic biomarkers. Molecular alterations that can predict meningioma behavior may be integrated into the upcoming World Health Organization grading system.
Figure
Reference
-
1. Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010; 99:307–14.
Article2. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro Oncol. 2018; 20:iv1–86.
Article3. Tufan K, Dogulu F, Kurt G, Emmez H, Ceviker N, Baykaner MK. Intracranial meningiomas of childhood and adolescence. Pediatr Neurosurg. 2005; 41:1–7.
Article4. Kotecha RS, Pascoe EM, Rushing EJ, et al. Meningiomas in children and adolescents: a meta-analysis of individual patient data. Lancet Oncol. 2011; 12:1229–39.
Article5. Kalamarides M, Stemmer-Rachamimov AO, Niwa-Kawakita M, et al. Identification of a progenitor cell of origin capable of generating diverse meningioma histological subtypes. Oncogene. 2011; 30:2333–44.
Article6. Preusser M, Brastianos PK, Mawrin C. Advances in meningioma genetics: novel therapeutic opportunities. Nat Rev Neurol. 2018; 14:106–15.
Article7. Galani V, Lampri E, Varouktsi A, Alexiou G, Mitselou A, Kyritsis AP. Genetic and epigenetic alterations in meningiomas. Clin Neurol Neurosurg. 2017; 158:119–25.
Article8. Sadetzki S, Flint-Richter P, Ben-Tal T, Nass D. Radiation-induced meningioma: a descriptive study of 253 cases. J Neurosurg. 2002; 97:1078–82.
Article9. Phillips LE, Frankenfeld CL, Drangsholt M, Koepsell TD, van Belle G, Longstreth WT. Intracranial meningioma and ionizing radiation in medical and occupational settings. Neurology. 2005; 64:350–2.
Article10. Schneider B, Pülhorn H, Röhrig B, Rainov NG. Predisposing conditions and risk factors for development of symptomatic meningioma in adults. Cancer Detect Prev. 2005; 29:440–7.
Article11. Flint-Richter P, Mandelzweig L, Oberman B, Sadetzki S. Possible interaction between ionizing radiation, smoking, and gender in the causation of meningioma. Neuro Oncol. 2011; 13:345–52.
Article12. Phillips LE, Koepsell TD, van Belle G, Kukull WA, Gehrels JA, Longstreth WT Jr. History of head trauma and risk of intracranial meningioma: population-based case-control study. Neurology. 2002; 58:1849–52.
Article13. Huisman TW, Tanghe HJ, Koper JW, et al. Progesterone, oestradiol, somatostatin and epidermal growth factor receptors on human meningiomas and their CT characteristics. European Journal of Cancer and Clinical Oncology. 1991; 27:1453–7.
Article14. Sioka C, Kyritsis AP. Chemotherapy, hormonal therapy, and immunotherapy for recurrent meningiomas. J Neurooncol. 2009; 92:1–6.
Article15. Suppiah S, Nassiri F, Bi WL, et al. Molecular and translational advances in meningiomas. Neuro Oncol. 2019; 21:i4–17.16. Simpson D. The recurrence of intracranial meningiomas after surgical treatment. J Neurol Neurosurg Psychiatry. 1957; 20:22–39.
Article17. Gousias K, Schramm J, Simon M. The Simpson grading revisited: aggressive surgery and its place in modern meningioma management. J Neurosurg. 2016; 125:551–60.
Article18. Yuzawa S, Nishihara H, Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016; 33:237–47.
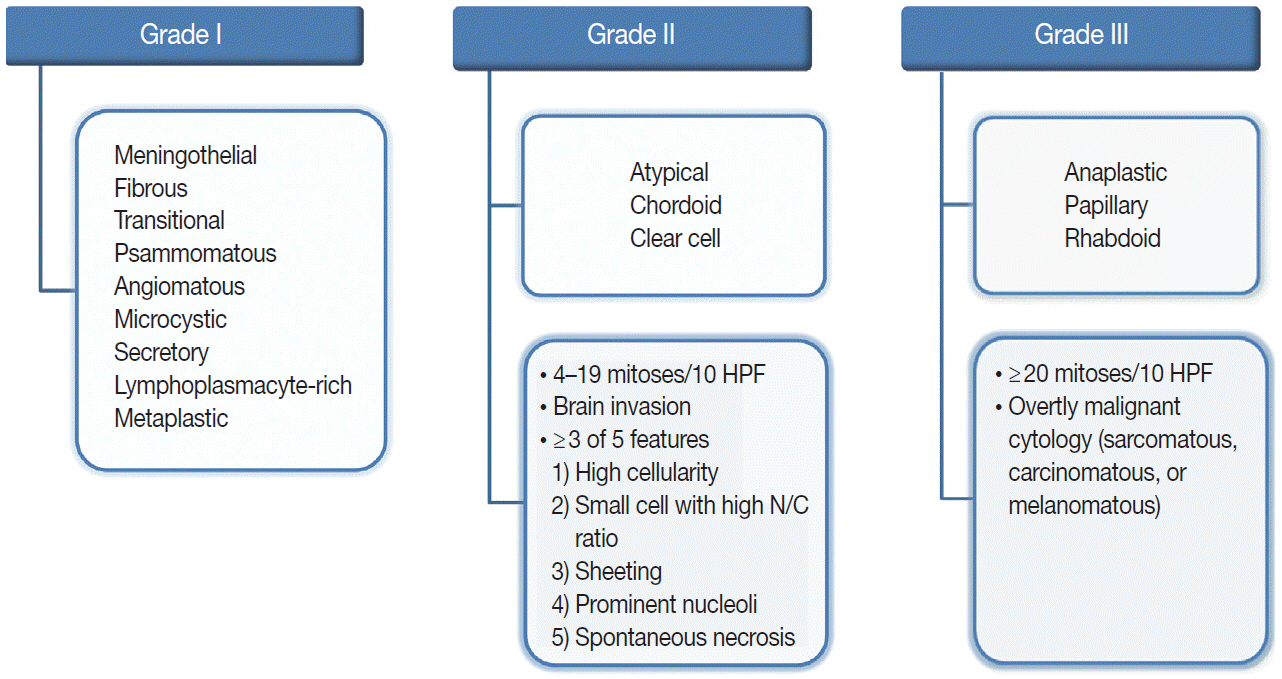
Article19. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO classification of tumours of the central nervous system. Revised 4th ed. Lyon: International Agency for Research on Cancer;2016.20. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016; 131:803–20.
Article21. Zankl H, Zang KD. Cytological and cytogenetical studies on brain tumors. 4. Identification of the missing G chromosome in human meningiomas as no. 22 by fluorescence technique. Humangenetik. 1972; 14:167–9.22. Proctor DT, Ramachandran S, Lama S, Sutherland GR. Towards molecular classification of meningioma: evolving treatment and diagnostic paradigms. World Neurosurg. 2018; 119:366–73.
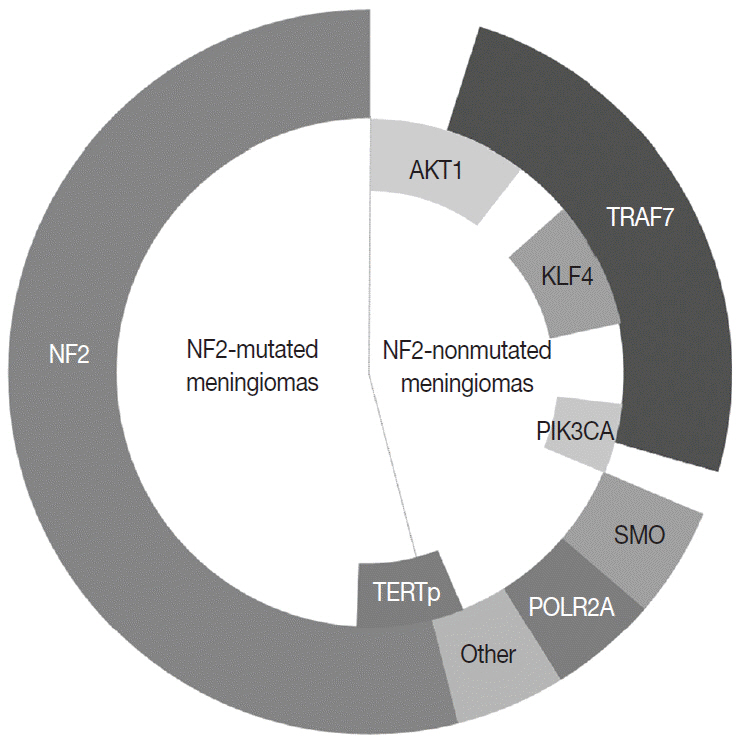
Article23. Ruttledge MH, Sarrazin J, Rangaratnam S, et al. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994; 6:180–4.24. Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013; 339:1077–80.25. Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013; 45:285–9.26. Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro Oncol. 2016; 18:649–55.27. Seizinger BR, de la Monte S, Atkins L, Gusella JF, Martuza RL. Molecular genetic approach to human meningioma: loss of genes on chromosome 22. Proc Natl Acad Sci U S A. 1987; 84:5419–23.
Article28. Mawrin C, Perry A. Pathological classification and molecular genetics of meningiomas. J Neurooncol. 2010; 99:379–91.
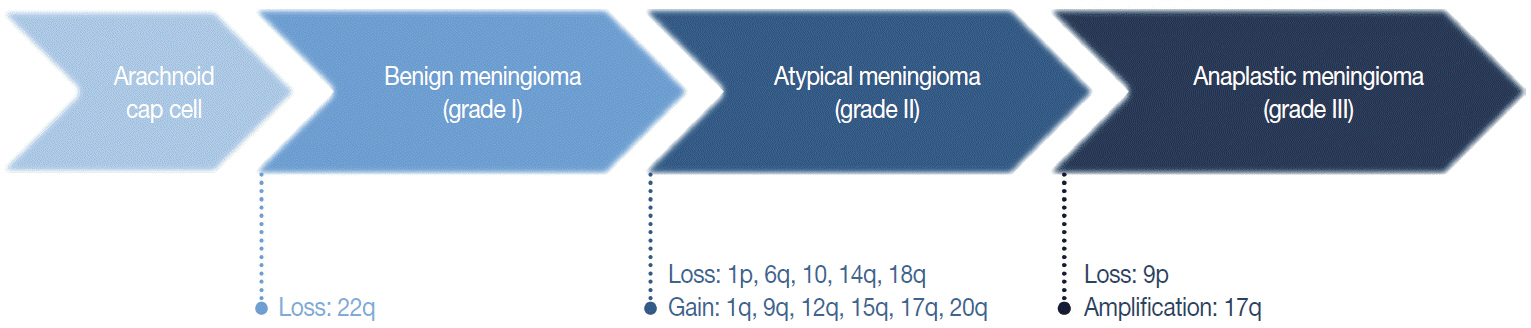
Article29. Weber RG, Boström J, Wolter M, et al. Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci U S A. 1997; 94:14719–24.
Article30. Abedalthagafi MS, Merrill PH, Bi WL, et al. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 2014; 5:10596–606.
Article31. Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro Oncol. 2016; 18:269–74.
Article32. Cai DX, Banerjee R, Scheithauer BW, Lohse CM, Kleinschmidt-Demasters BK, Perry A. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001; 60:628–36.
Article33. Lamszus K. Meningioma pathology, genetics, and biology. J Neuropathol Exp Neurol. 2004; 63:275–86.
Article34. Lomas J, Bello MJ, Arjona D, et al. Analysis of p73 gene in meningiomas with deletion at 1p. Cancer Genet Cytogenet. 2001; 129:88–91.
Article35. Bostrom J, Meyer-Puttlitz B, Wolter M, et al. Alterations of the tumor suppressor genes CDKN2A (p16(INK4a)), p14(ARF), CDKN2B (p15(INK4b)), and CDKN2C (p18(INK4c)) in atypical and anaplastic meningiomas. Am J Pathol. 2001; 159:661–9.36. Mendiola M, Bello MJ, Alonso J, et al. Search for mutations of the hRAD54 gene in sporadic meningiomas with deletion at 1p32. Mol Carcinog. 1999; 24:300–4.37. Piaskowski S, Rieske P, Szybka M, et al. GADD45A and EPB41 as tumor suppressor genes in meningioma pathogenesis. Cancer Genet Cytogenet. 2005; 162:63–7.38. Lusis EA, Watson MA, Chicoine MR, et al. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Res. 2005; 65:7121–6.39. Zhang X, Gejman R, Mahta A, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010; 70:2350–8.
Article40. Och W, Szmuda T, Sikorska B, et al. Recurrence-associated chromosomal anomalies in meningiomas: Single-institution study and a systematic review with meta-analysis. Neurol Neurochir Pol. 2016; 50:439–48.
Article41. Perry A, Banerjee R, Lohse CM, Kleinschmidt-DeMasters BK, Scheithauer BW. A role for chromosome 9p21 deletions in the malignant progression of meningiomas and the prognosis of anaplastic meningiomas. Brain Pathol. 2002; 12:183–90.42. Buschges R, Ichimura K, Weber RG, Reifenberger G, Collins VP. Allelic gain and amplification on the long arm of chromosome 17 in anaplastic meningiomas. Brain Pathol. 2002; 12:145–53.43. Smith MJ. Germline and somatic mutations in meningiomas. Cancer Genet. 2015; 208:107–14.
Article44. Kerr K, Qualmann K, Esquenazi Y, Hagan J, Kim DH. Familial syndromes involving meningiomas provide mechanistic insight Into sporadic disease. Neurosurgery. 2018; 83:1107–18.
Article45. Smith MJ, Higgs JE, Bowers NL, et al. Cranial meningiomas in 411 neurofibromatosis type 2 (NF2) patients with proven gene mutations: clear positional effect of mutations, but absence of female severity effect on age at onset. J Med Genet. 2011; 48:261–5.
Article46. Antinheimo J, Haapasalo H, Haltia M, et al. Proliferation potential and histological features in neurofibromatosis 2-associated and sporadic meningiomas. J Neurosurg. 1997; 87:610–4.
Article47. Christiaans I, Kenter SB, Brink HC, et al. Germline SMARCB1 mutation and somatic NF2 mutations in familial multiple meningiomas. J Med Genet. 2011; 48:93–7.48. Smith MJ, O'Sullivan J, Bhaskar SS, et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nat Genet. 2013; 45:295–8.49. Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999; 59:74–9.50. Hulsebos TJ, Plomp AS, Wolterman RA, Robanus-Maandag EC, Baas F, Wesseling P. Germline mutation of INI1/SMARCB1 in familial schwannomatosis. Am J Hum Genet. 2007; 80:805–10.51. Tsurusaki Y, Okamoto N, Ohashi H, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012; 44:376–8.52. Smith MJ, Wallace AJ, Bowers NL, et al. Frequency of SMARCB1 mutations in familial and sporadic schwannomatosis. Neurogenetics. 2012; 13:141–5.53. Smith MJ, Wallace AJ, Bowers NL, Eaton H, Evans DG. SMARCB1 mutations in schwannomatosis and genotype correlations with rhabdoid tumors. Cancer Genet. 2014; 207:373–8.54. Schmitz U, Mueller W, Weber M, Sevenet N, Delattre O, von Deimling A. INI1 mutations in meningiomas at a potential hotspot in exon 9. Br J Cancer. 2001; 84:199–201.55. Smith MJ, Wallace AJ, Bennett C, et al. Germline SMARCE1 mutations predispose to both spinal and cranial clear cell meningiomas. J Pathol. 2014; 234:436–40.56. Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017; 19:535–45.57. Haugh AM, Njauw CN, Bubley JA, et al. Genotypic and phenotypic features of BAP1 cancer syndrome: a report of 8 new families and review of cases in the literature. JAMA Dermatol. 2017; 153:999–1006.58. Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene. 1999; 18:7844–51.
Article59. Smith MJ, Beetz C, Williams SG, et al. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol. 2014; 32:4155–61.60. Ng JM, Curran T. The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011; 11:493–501.
Article61. Aavikko M, Li SP, Saarinen S, et al. Loss of SUFU function in familial multiple meningioma. Am J Hum Genet. 2012; 91:520–6.
Article62. Choy W, Kim W, Nagasawa D, et al. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurg Focus. 2011; 30:E6.
Article63. Nunes F, Shen Y, Niida Y, et al. Inactivation patterns of NF2 and DAL-1/4.1B (EPB41L3) in sporadic meningioma. Cancer Genet Cytogenet. 2005; 162:135–9.64. Pec´ina-Šlaus N. Merlin, the NF2 gene product. Pathol Oncol Res. 2013; 19:365–73.65. Lallemand D, Manent J, Couvelard A, et al. Merlin regulates transmembrane receptor accumulation and signaling at the plasma membrane in primary mouse Schwann cells and in human schwannomas. Oncogene. 2009; 28:854–65.
Article66. Goutagny S, Yang HW, Zucman-Rossi J, et al. Genomic profiling reveals alternative genetic pathways of meningioma malignant progression dependent on the underlying NF2 status. Clin Cancer Res. 2010; 16:4155–64.
Article67. Zotti T, Scudiero I, Vito P, Stilo R. The emerging role of TRAF7 in tumor development. J Cell Physiol. 2017; 232:1233–8.68. Bouwmeester T, Bauch A, Ruffner H, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004; 6:97–105.69. Xu LG, Li LY, Shu HB. TRAF7 potentiates MEKK3-induced AP1 and CHOP activation and induces apoptosis. J Biol Chem. 2004; 279:17278–82.70. Reuss DE, Piro RM, Jones DT, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013; 125:351–8.71. Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nat Rev Cancer. 2013; 13:701–13.72. Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000; 275:28230–9.73. Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007; 448:439–44.74. Yesiloz U, Kirches E, Hartmann C, et al. Frequent AKT1E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro Oncol. 2017; 19:1088–96.75. Keppler-Noreuil KM, Baker EH, Sapp JC, Lindhurst MJ, Biesecker LG. Somatic AKT1 mutations cause meningiomas colocalizing with a characteristic pattern of cranial hyperostosis. Am J Med Genet A. 2016; 170:2605–10.76. Boetto J, Bielle F, Sanson M, Peyre M, Kalamarides M. SMO mutation status defines a distinct and frequent molecular subgroup in olfactory groove meningiomas. Neuro Oncol. 2017; 19:345–51.77. Strickland MR, Gill CM, Nayyar N, et al. Targeted sequencing of SMO and AKT1 in anterior skull base meningiomas. J Neurosurg. 2017; 127:438–44.78. Zadeh G, Karimi S, Aldape KD. PIK3CA mutations in meningioma. Neuro Oncol. 2016; 18:603–4.79. Clark VE, Harmanci AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016; 48:1253–9.
Article80. Poulikakos PI, Xiao GH, Gallagher R, Jablonski S, Jhanwar SC, Testa JR. Re-expression of the tumor suppressor NF2/merlin inhibits invasiveness in mesothelioma cells and negatively regulates FAK. Oncogene. 2006; 25:5960–8.
Article81. Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014; 14:598–610.
Article82. Wang X, Gong Y, Wang D, et al. Analysis of gene expression profiling in meningioma: deregulated signaling pathways associated with meningioma and EGFL6 overexpression in benign meningioma tissue and serum. PLoS One. 2012; 7:e52707.
Article83. Murali R, Wiesner T, Scolyer RA. Tumours associated with BAP1 mutations. Pathology. 2013; 45:116–26.84. Shankar GM, Santagata S. BAP1 mutations in high-grade meningioma: implications for patient care. Neuro Oncol. 2017; 19:1447–56.85. Harmanci AS, Youngblood MW, Clark VE, et al. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017; 8:14433.
Article86. Evans LT, Van Hoff J, Hickey WF, et al. SMARCE1 mutations in pediatric clear cell meningioma: case report. J Neurosurg Pediatr. 2015; 16:296–300.87. Loo E, Khalili P, Beuhler K, Siddiqi I, Vasef MA. BRAF V600E mutation across multiple tumor types: correlation between DNA-based sequencing and mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol. 2018; 26:709–13.88. Bujko M, Kober P, Tysarowski A, et al. EGFR, PIK3CA, KRAS and BRAF mutations in meningiomas. Oncol Lett. 2014; 7:2019–22.89. Mordechai O, Postovsky S, Vlodavsky E, et al. Metastatic rhabdoid meningioma with BRAF V600E mutation and good response to personalized therapy: case report and review of the literature. Pediatr Hematol Oncol. 2015; 32:207–11.90. Behling F, Barrantes-Freer A, Skardelly M, et al. Frequency of BRAF V600E mutations in 969 central nervous system neoplasms. Diagn Pathol. 2016; 11:55.
Article91. Baia GS, Stifani S, Kimura ET, McDermott MW, Pieper RO, Lal A. Notch activation is associated with tetraploidy and enhanced chromosomal instability in meningiomas. Neoplasia. 2008; 10:604–12.
Article92. Yang HW, Kim TM, Song SS, et al. Alternative splicing of CHEK2 and codeletion with NF2 promote chromosomal instability in meningioma. Neoplasia. 2012; 14:20–8.93. Lino MM, Merlo A. PI3Kinase signaling in glioblastoma. J Neurooncol. 2011; 103:417–27.
Article94. Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003; 11:11–23.
Article95. Peters N, Wellenreuther R, Rollbrocker B, et al. Analysis of the PTEN gene in human meningiomas. Neuropathol Appl Neurobiol. 1998; 24:3–8.96. Juratli TA, McCabe D, Nayyar N, et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 2018; 136:779–92.
Article97. Koelsche C, Sahm F, Capper D, et al. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013; 126:907–15.98. Gao K, Li G, Qu Y, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016; 7:8712–25.99. Simon M, Park TW, Leuenroth S, Hans VH, Loning T, Schramm J. Telomerase activity and expression of the telomerase catalytic subunit, hTERT, in meningioma progression. J Neurosurg. 2000; 92:832–40.
Article100. Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2016; 108:djv377.101. Goutagny S, Nault JC, Mallet M, Henin D, Rossi JZ, Kalamarides M. High incidence of activating TERT promoter mutations in meningiomas undergoing malignant progression. Brain Pathol. 2014; 24:184–9.102. He S, Pham MH, Pease M, et al. A review of epigenetic and gene expression alterations associated with intracranial meningiomas. Neurosurg Focus. 2013; 35:E5.
Article103. Olar A, Wani KM, Wilson CD, et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017; 133:431–44.
Article104. Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017; 18:682–94.105. Nassiri F, Mamatjan Y, Suppiah S, et al. DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019; Jun. 3. [Epub]. https://doi.org/10.1093/neuonc/noz061.
Article106. Liu Y, Pang JC, Dong S, Mao B, Poon WS, Ng HK. Aberrant CpG island hypermethylation profile is associated with atypical and anaplastic meningiomas. Hum Pathol. 2005; 36:416–25.
Article107. Pereira BJ, Oba-Shinjo SM, de Almeida AN, Marie SK. Molecular alterations in meningiomas: Literature review. Clin Neurol Neurosurg. 2019; 176:89–96.
Article108. Barski D, Wolter M, Reifenberger G, Riemenschneider MJ. Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathol. 2010; 20:623–31.109. Bello MJ, Amiñoso C, Lopez-Marin I, et al. DNA methylation of multiple promoter-associated CpG islands in meningiomas: relationship with the allelic status at 1p and 22q. Acta Neuropathol. 2004; 108:413–21.
Article110. Nakane Y, Natsume A, Wakabayashi T, et al. Malignant transformation-related genes in meningiomas: allelic loss on 1p36 and methylation status of p73 and RASSF1A. J Neurosurg. 2007; 107:398–404.
Article111. Zhou Y, Zhong Y, Wang Y, et al. Activation of p53 by MEG3 noncoding RNA. J Biol Chem. 2007; 282:24731–42.112. Kishida Y, Natsume A, Kondo Y, et al. Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis. 2012; 33:436–41.
Article113. Jun P, Hong C, Lal A, et al. Epigenetic silencing of the kinase tumor suppressor WNK2 is tumor-type and tumor-grade specific. Neuro Oncol. 2009; 11:414–22.114. Gao F, Shi L, Russin J, et al. DNA methylation in the malignant transformation of meningiomas. PLoS One. 2013; 8:e54114.
Article115. Nair SS, Kumar R. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol Oncol. 2012; 6:611–9.
Article116. Kumar R, Li DQ, Muller S, Knapp S. Epigenomic regulation of oncogenesis by chromatin remodeling. Oncogene. 2016; 35:4423–36.
Article117. Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016; 17:284–99.
Article118. Murnyák B, Bognár L, Klekner Á, Hortobágyi T. Epigenetics of meningiomas. Biomed Res Int. 2015; 2015:532451.
Article119. Katz LM, Hielscher T, Liechty B, et al. Loss of histone H3K27me3 identifies a subset of meningiomas with increased risk of recurrence. Acta Neuropathol. 2018; 135:955–63.
Article120. Pérez-Magán E, Rodríguez de Lope A, Ribalta T, et al. Differential expression profiling analyses identifies downregulation of 1p, 6q, and 14q genes and overexpression of 6p histone cluster 1 genes as markers of recurrence in meningiomas. Neuro Oncol. 2010; 12:1278–90.
Article121. Lu C, Allis CD. SWI/SNF complex in cancer. Nat Genet. 2017; 49:178–9.
Article122. Collord G, Tarpey P, Kurbatova N, et al. An integrated genomic analysis of anaplastic meningioma identifies prognostic molecular signatures. Sci Rep. 2018; 8:13537.
Article123. Zhi F, Zhou G, Wang S, et al. A microRNA expression signature predicts meningioma recurrence. Int J Cancer. 2013; 132:128–36.
Article124. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009; 4:199–227.
Article125. Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004; 101:2999–3004.
Article126. Venza M, Visalli M, Beninati C, et al. Involvement of epimutations in meningioma. Brain Tumor Pathol. 2015; 32:163–8.
Article127. Kliese N, Gobrecht P, Pachow D, et al. miRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013; 32:4712–20.
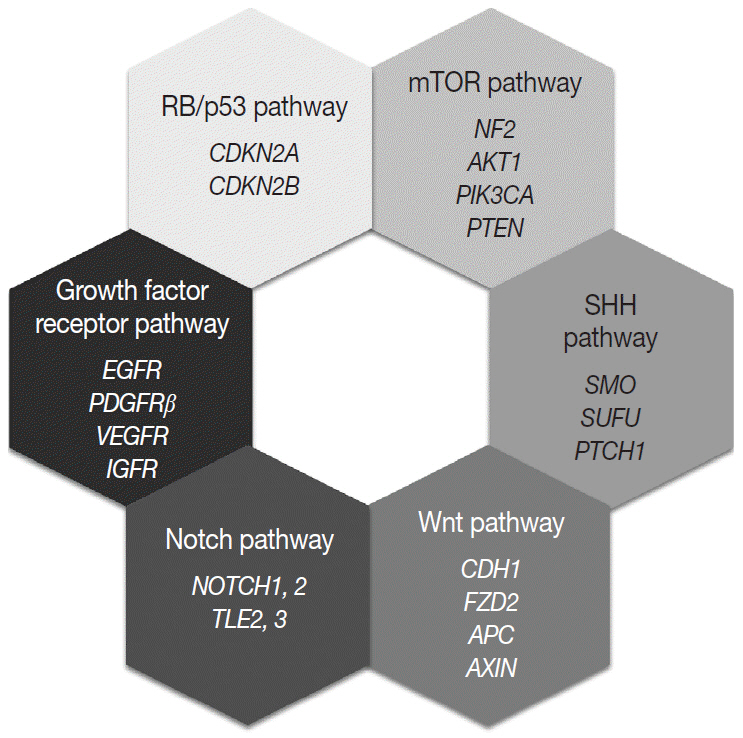
Article128. Domingues P, González-Tablas M, Otero Á, et al. Genetic/molecular alterations of meningiomas and the signaling pathways targeted. Oncotarget. 2015; 6:10671–88.
Article129. Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR signaling in cancer. Front Oncol. 2014; 4:64.
Article130. Sato T, Sekido Y. NF2/Merlin inactivation and potential therapeutic targets in mesothelioma. Int J Mol Sci. 2018; 19:E988.
Article131. Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017; 168:960–76.
Article132. Beltrami S, Kim R, Gordon J. Neurofibromatosis type 2 protein, NF2: an uncoventional cell cycle regulator. Anticancer Res. 2013; 33:1–11.133. Rimkus TK, Carpenter RL, Qasem S, Chan M, Lo HW. Targeting the sonic Hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel). 2016; 8:E22.
Article134. Nigim F, Wakimoto H, Kasper EM, Ackermans L, Temel Y. Emerging medical treatments for meningioma in the molecular era. Biomedicines. 2018; 6:E86.
Article135. Miller R Jr, DeCandio ML, Dixon-Mah Y, et al. Molecular targets and treatment of meningioma. J Neurol Neurosurg. 2014; 1:1000101.
Article136. Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004; 303:1483–7.137. Ragel BT, Jensen RL. Aberrant signaling pathways in meningiomas. J Neurooncol. 2010; 99:315–24.
Article138. Kros J, de Greve K, van Tilborg A, et al. NF2 status of meningiomas is associated with tumour localization and histology. J Pathol. 2001; 194:367–72.
Article