Cancer Res Treat.
2020 Apr;52(2):492-504. 10.4143/crt.2019.457.
EBV-miR-BHRF1-1 Targets p53 Gene: Potential Role in Epstein-BarrVirus Associated Chronic Lymphocytic Leukemia
- Affiliations
-
- 1Department of Hematology, The First Affiliated Hospital of Nanjing Medical University, Jiangsu Province Hospital, Nanjing, China
- 2Key Laboratory of Hematology of Nanjing Medical University, Nanjing, China
- 3Collaborative Innovation Center for Cancer Personalized Medicine, Nanjing, China
- KMID: 2500334
- DOI: http://doi.org/10.4143/crt.2019.457
Abstract
- Purpose
The purpose of this study was to investigate the prognostic impact of Epstein-Barr virus (EBV)–microRNA (miRNA, miR)-BHRF1-1 with chronic lymphocytic leukemia (CLL) as well as role of EBV-miR-BHRF1-1 in p53 gene.
Materials and Methods
Quantitative reverse transcription–polymerase chain reaction and western blotting were used to quantify EBV-miR-BHRF1-1 and p53 expression in cultured CLL.
Results
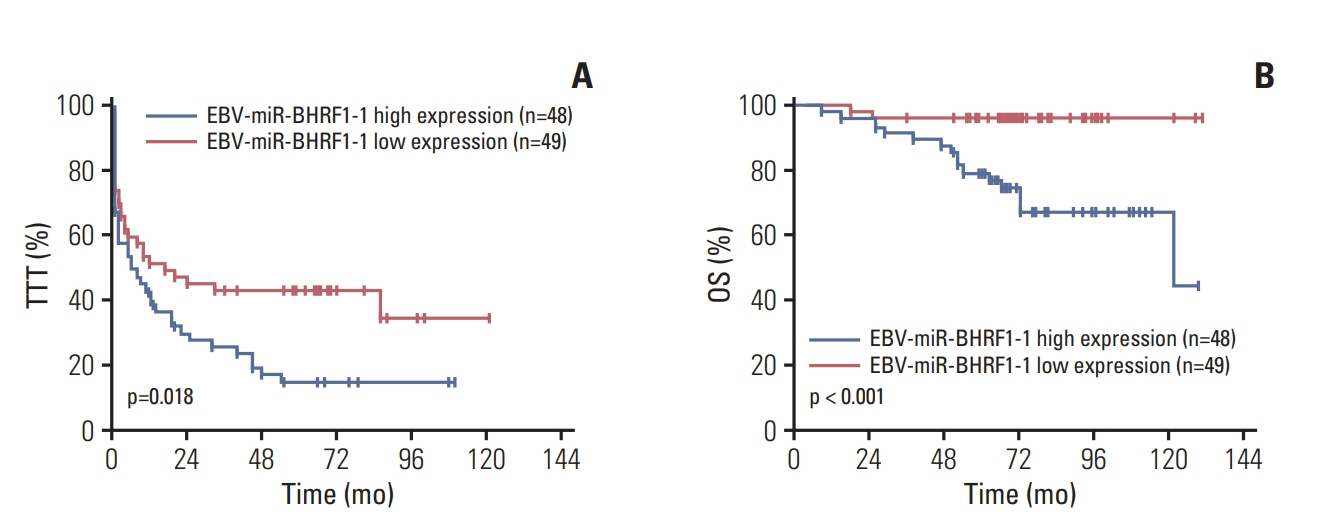
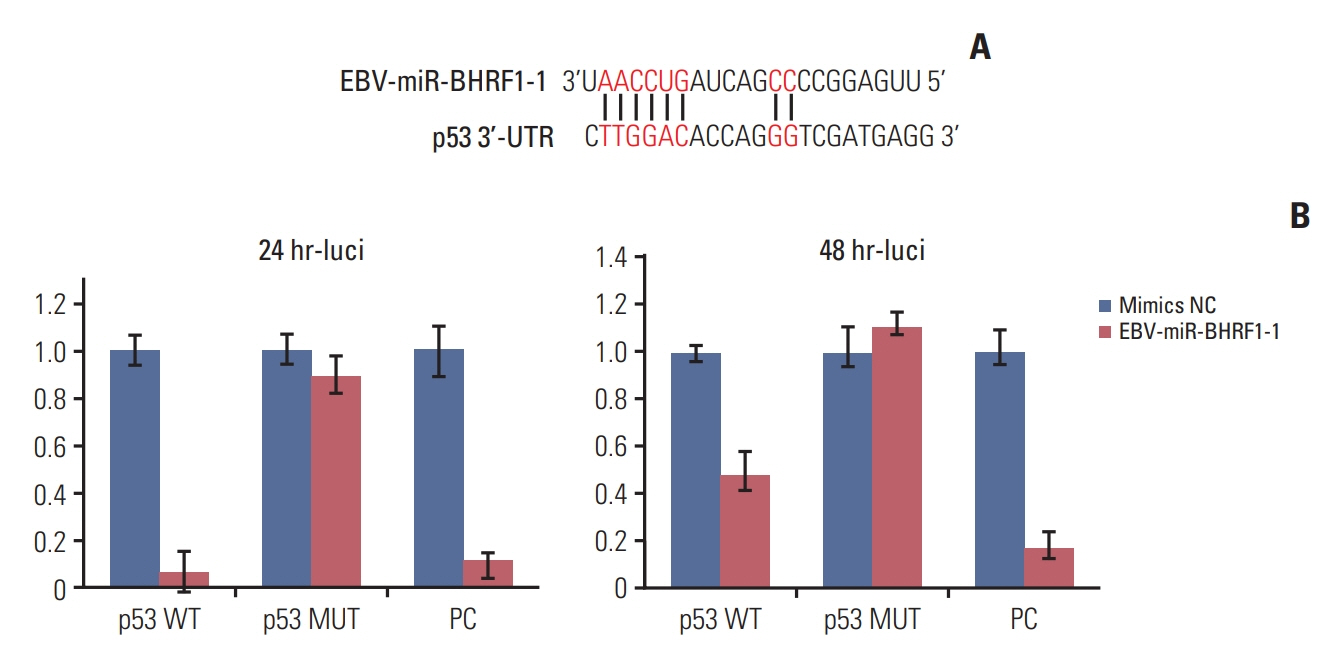
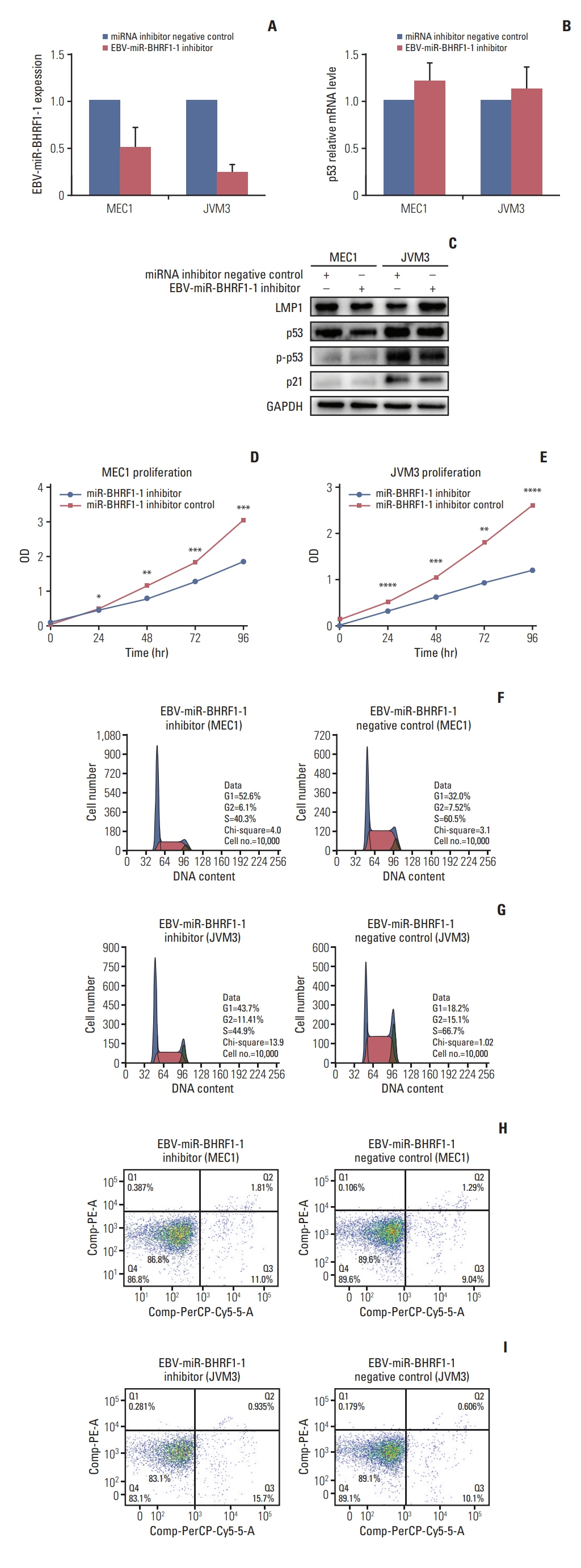
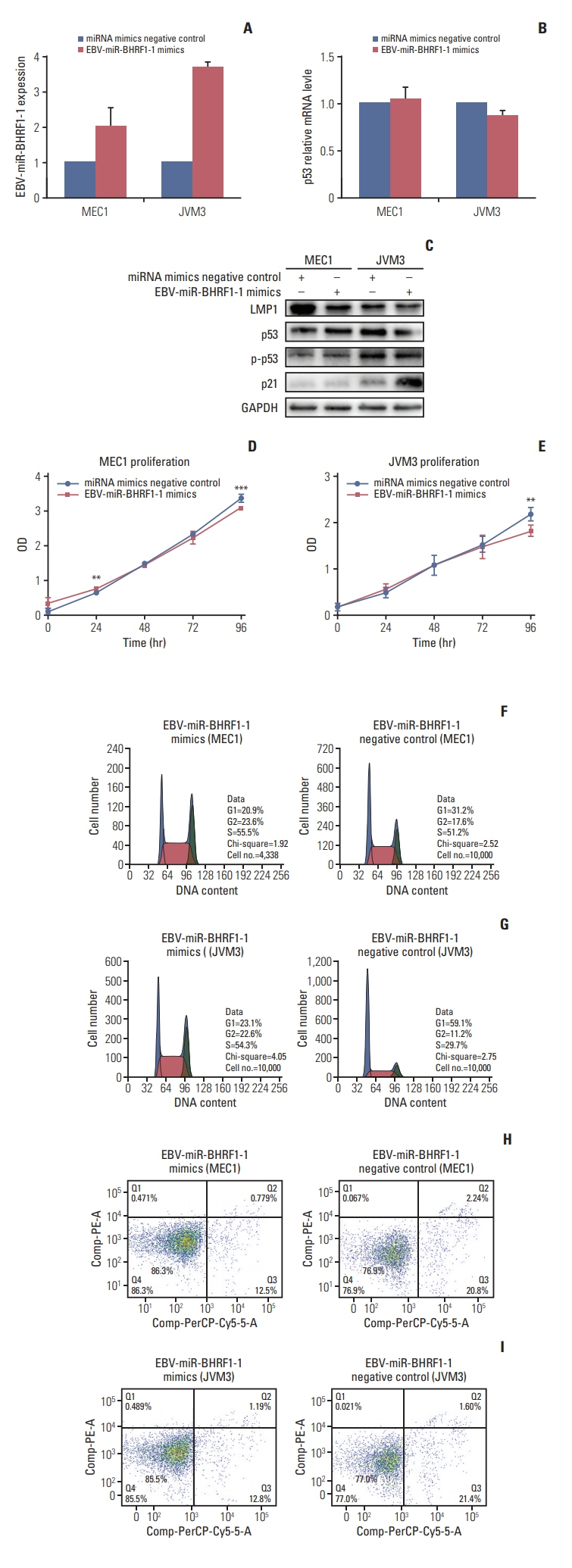
p53 aberration was associated with the higher expression level of EBV-miR-BHRF1-1 (p < 0.001) which was also an independent prognostic marker for overall survival (p=0.028; hazard ratio, 5.335; 95% confidence interval, 1.193 to 23.846) in 97 newly-diagnosed CLL patients after adjusted with International Prognostic Index for patients with CLL. We identified EBV-miR-BHRF1-1 as a viral miRNA regulator of p53. EBV-miR-BHRF1-1 repressed luciferase reporter activity by specific interaction with the seed region within the p53 3- untranslated region. Discordance of p53 messenger RNA and protein expression was associated with high EBV-miR-BHRF1-1 levels in CLL patients and cell lines. EBV-miR-BHRF1- 1 inhibition upregulated p53 protein expression, induced cell cycle arrest and apoptosis and decreased cell proliferation in cell lines. EBV-miR-BHRF1-1 mimics downregulated p53 protein expression, decreased cell cycle arrest and apoptosis, and induced cell proliferation in cell lines.
Conclusion
This study supported the role of EBV-miR-BHRF1-1 in p53 regulation in vitro. Our results support the potential of EBV-miR-BHRF1-1 as a therapeutic target in EBV-associated CLL with p53 gene aberration.
Keyword
Figure
Reference
-
References
1. Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, et al. Identification of virus-encoded microRNAs. Science. 2004; 304:734–6.
Article2. Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, et al. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011; 7:e1002193.
Article3. Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, et al. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011; 39:1880–93.
Article4. Barth S, Meister G, Grasser FA. EBV-encoded miRNAs. Biochim Biophys Acta. 2011; 1809:631–40.
Article5. Navari M, Etebari M, Ibrahimi M, Leoncini L, Piccaluga PP. Pathobiologic roles of Epstein-Barr virus-encoded microRNAs in human lymphomas. Int J Mol Sci. 2018; 19:E1168.
Article6. Liang JH, Gao R, Xia Y, Gale RP, Chen RZ, Yang YQ, et al. Prognostic impact of Epstein-Barr virus (EBV)-DNA copy number at diagnosis in chronic lymphocytic leukemia. Oncotarget. 2016; 7:2135–42.
Article7. Visco C, Falisi E, Young KH, Pascarella M, Perbellini O, Carli G, et al. Epstein-Barr virus DNA load in chronic lymphocytic leukemia is an independent predictor of clinical course and survival. Oncotarget. 2015; 6:18653–63.
Article8. Baliakas P, Hadzidimitriou A, Sutton LA, Rossi D, Minga E, Villamor N, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015; 29:329–36.
Article9. Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med. 2016; 374:311–22.
Article10. Burger JA, Sivina M, Jain N, Kim E, Kadia T, Estrov Z, et al. Randomized trial of ibrutinib vs ibrutinib plus rituximab in patients with chronic lymphocytic leukemia. Blood. 2019; 133:1011–9.
Article11. Li Z, Chen X, Li L, Liu S, Yang L, Ma X, et al. EBV encoded miR-BHRF1-1 potentiates viral lytic replication by downregulating host p53 in nasopharyngeal carcinoma. Int J Biochem Cell Biol. 2012; 44:275–9.
Article12. Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008; 111:5446–56.
Article13. International CLL-IPI Working Group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016; 17:779–90.14. Tarrand JJ, Keating MJ, Tsimberidou AM, O'Brien S, LaSala RP, Han XY, et al. Epstein-Barr virus latent membrane protein 1 mRNA is expressed in a significant proportion of patients with chronic lymphocytic leukemia. Cancer. 2010; 116:880–7.
Article15. Renouf B, Hollville E, Pujals A, Tetaud C, Garibal J, Wiels J. Activation of p53 by MDM2 antagonists has differential apoptotic effects on Epstein-Barr virus (EBV)-positive and EBVnegative Burkitt's lymphoma cells. Leukemia. 2009; 23:1557–63.
Article16. AlQarni S, Al-Sheikh Y, Campbell D, Drotar M, Hannigan A, Boyle S, et al. Lymphomas driven by Epstein-Barr virus nuclear antigen-1 (EBNA1) are dependant upon Mdm2. Oncogene. 2018; 37:3998–4012.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Epstein-Barr Virus and p53 in Laryngeal and Nasopharyngeal Carcinomas
- The role of promoter methylation in Epstein-Barr virus (EBV) microRNA expression in EBV-infected B cell lines
- Immunohistochemical and SSCP analysis of p53 in malignant lymphomas
- Correlation between Expression of p53 and Bcl-2 Protein and Epstein-Barr Virus Detection in Gastric Adenocarcinoma
- Regulation of microRNA-7-5p and LRP6 by Epstein-Barr Virus-Encoded RNAs in Burkitt's Lymphoma Cell Line Akata